staufen double-stranded RNA binding protein 2
ZFIN 
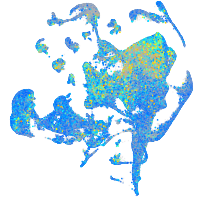
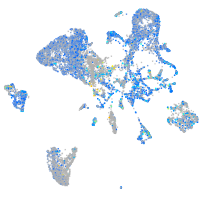
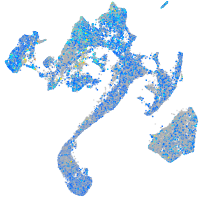
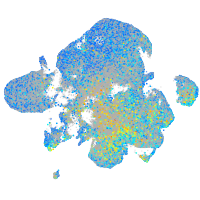
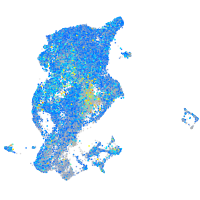
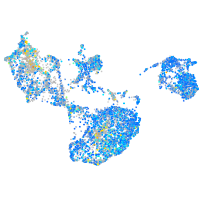
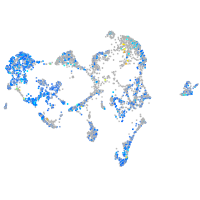
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kif5bb | 0.143 | si:dkey-151g10.6 | -0.105 |
| rab2a | 0.142 | hmgb2b | -0.101 |
| atp1a1a.1 | 0.142 | snrpf | -0.100 |
| rap1gap2a | 0.140 | her15.1 | -0.098 |
| scg2a | 0.140 | apex1 | -0.097 |
| jund | 0.140 | ybx1 | -0.089 |
| calm1a | 0.137 | stmn1a | -0.087 |
| tpi1b | 0.135 | CU467822.1 | -0.087 |
| fxyd1 | 0.133 | snrpe | -0.087 |
| msi2b | 0.132 | eif4a1a | -0.085 |
| mt-co1 | 0.132 | her4.2 | -0.085 |
| CU929259.1 | 0.131 | rrm1 | -0.083 |
| vapb | 0.131 | rplp1 | -0.080 |
| atp1b1a | 0.128 | ddx39ab | -0.080 |
| gapdhs | 0.128 | pcna | -0.079 |
| vamp2 | 0.128 | XLOC-003692 | -0.078 |
| cst3 | 0.126 | rps28 | -0.078 |
| tmem176l.4 | 0.126 | sae1 | -0.078 |
| eno1a | 0.125 | rpl39 | -0.076 |
| syt1a | 0.124 | neurod4 | -0.076 |
| gnb1b | 0.123 | rpl36a | -0.076 |
| ptbp3 | 0.122 | esco2 | -0.075 |
| mbnl2 | 0.122 | hmga1a | -0.075 |
| pkma | 0.122 | hnrnpabb | -0.075 |
| cldnh | 0.119 | npm1a | -0.075 |
| akap10 | 0.119 | rpl38 | -0.075 |
| b2ml | 0.118 | dut | -0.073 |
| aldocb | 0.118 | uba2 | -0.073 |
| flot2a | 0.117 | cbx3a | -0.073 |
| slc4a10a | 0.117 | cdca5 | -0.073 |
| limch1b | 0.117 | rps23 | -0.072 |
| h1f0 | 0.117 | rps21 | -0.072 |
| mical3a | 0.117 | tk1 | -0.072 |
| CABZ01055365.1 | 0.116 | h3f3a | -0.072 |
| tspan3a | 0.116 | ccna2 | -0.071 |