stromal antigen 2a
ZFIN 
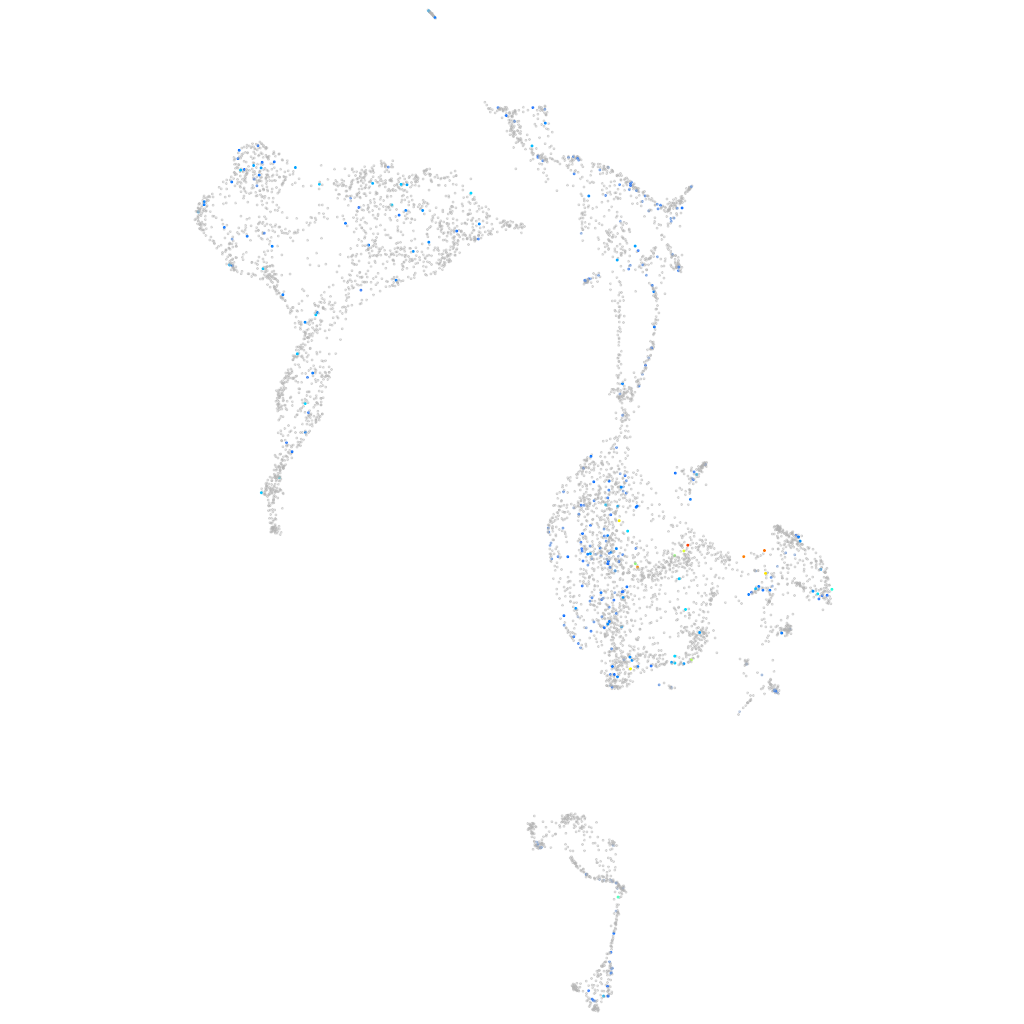
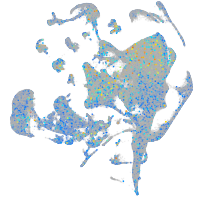
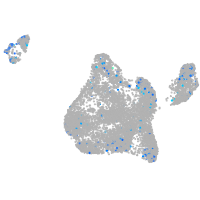
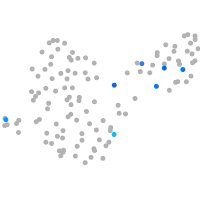
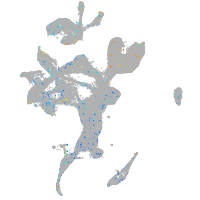
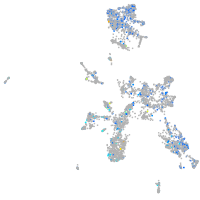
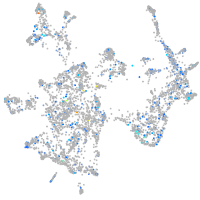
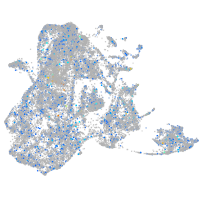
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| aldoca | 0.201 | hmga1a | -0.061 |
| BX649300.2 | 0.181 | syncrip | -0.054 |
| XLOC-043685 | 0.162 | lsm7 | -0.052 |
| lhfpl4b | 0.155 | hmgn6 | -0.052 |
| myot | 0.153 | hspb1 | -0.052 |
| pfkma | 0.148 | npm1a | -0.051 |
| ugt1a7 | 0.145 | cd2bp2 | -0.050 |
| lrit1a | 0.138 | fbl | -0.050 |
| zgc:195001 | 0.138 | bms1 | -0.050 |
| cdk5r2b | 0.136 | dkc1 | -0.050 |
| CABZ01058452.1 | 0.133 | rpl5b | -0.049 |
| tulp1b | 0.130 | hnrnpa1b | -0.049 |
| XLOC-005727 | 0.129 | rrs1 | -0.048 |
| CR855277.2 | 0.126 | eif2s2 | -0.048 |
| atf5a | 0.126 | nop58 | -0.048 |
| ucp3 | 0.122 | rsl1d1 | -0.048 |
| mttp | 0.118 | nop2 | -0.047 |
| dlx4b | 0.117 | marcksb | -0.047 |
| CT027832.1 | 0.116 | rpl7l1 | -0.047 |
| si:ch1073-100f3.2 | 0.115 | hnrnpa0b | -0.046 |
| BX323861.3 | 0.113 | si:ch73-281n10.2 | -0.046 |
| tfap2d | 0.107 | krr1 | -0.046 |
| pnp4b | 0.106 | pes | -0.046 |
| si:ch211-234h8.7 | 0.106 | insb | -0.046 |
| tapbpl | 0.103 | ptbp1b | -0.045 |
| slc6a17 | 0.103 | snrpe | -0.045 |
| gba2 | 0.103 | ncl | -0.045 |
| CABZ01072309.1 | 0.102 | nono | -0.044 |
| LOC795897 | 0.102 | srsf11 | -0.044 |
| rims1a | 0.101 | psmb1 | -0.044 |
| opn6b | 0.101 | UTP14C | -0.044 |
| zfpm2a | 0.101 | hdgfl2 | -0.043 |
| otpa | 0.100 | lyar | -0.043 |
| LOC103911545 | 0.098 | rbm17 | -0.043 |
| si:ch211-188f17.1 | 0.098 | hspe1 | -0.043 |