"ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1"
ZFIN 
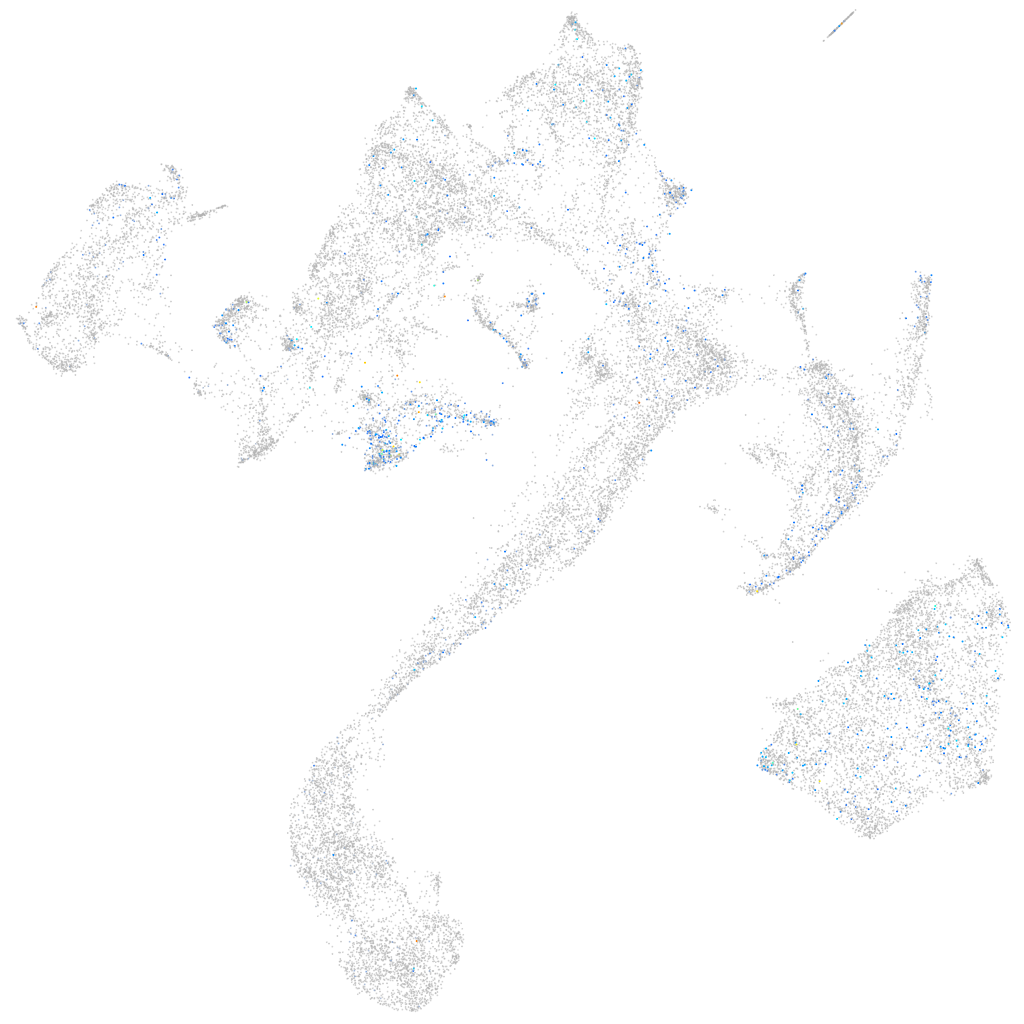
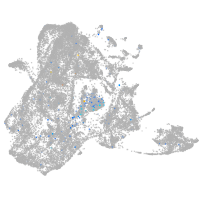
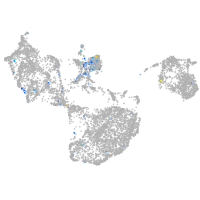
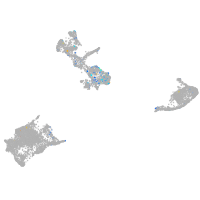
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.124 | XLOC-025819 | -0.053 |
| myt1b | 0.121 | nme2b.2 | -0.053 |
| tuba1c | 0.119 | gatm | -0.053 |
| nova2 | 0.115 | gamt | -0.052 |
| gpm6aa | 0.115 | pabpc4 | -0.052 |
| stmn1b | 0.107 | atp1a2a | -0.052 |
| tubb5 | 0.105 | si:ch73-367p23.2 | -0.052 |
| rnasekb | 0.100 | neb | -0.051 |
| rtn1b | 0.099 | ryr1b | -0.051 |
| cadm3 | 0.096 | XLOC-006515 | -0.051 |
| mmp25b | 0.095 | aldoab | -0.051 |
| CU467822.1 | 0.095 | XLOC-005350 | -0.051 |
| cspg5a | 0.094 | si:ch211-266g18.10 | -0.050 |
| gnao1a | 0.093 | ak1 | -0.050 |
| mdkb | 0.093 | ttn.1 | -0.049 |
| ckbb | 0.092 | actc1b | -0.049 |
| si:ch211-150g13.3 | 0.092 | actn3a | -0.049 |
| ptprjb.1 | 0.091 | ldb3b | -0.049 |
| elavl4 | 0.091 | atp2a1 | -0.049 |
| gng3 | 0.091 | pgam2 | -0.049 |
| nsg2 | 0.090 | eno3 | -0.049 |
| tuba1a | 0.090 | si:ch211-255p10.3 | -0.049 |
| sncb | 0.089 | prr33 | -0.049 |
| LOC100535907 | 0.089 | tmod4 | -0.048 |
| hmgb1b | 0.089 | XLOC-001975 | -0.048 |
| fam117ba | 0.089 | mybphb | -0.048 |
| sv2a | 0.089 | casq1b | -0.048 |
| ptmab | 0.088 | prx | -0.048 |
| scrt2 | 0.088 | tnnc2 | -0.048 |
| atpv0e2 | 0.087 | jph2 | -0.047 |
| hmgb3a | 0.086 | eef2l2 | -0.047 |
| stmn2a | 0.086 | CR855257.1 | -0.047 |
| zgc:165603 | 0.086 | myom2a | -0.047 |
| tubb2b | 0.086 | ckmb | -0.047 |
| syt2a | 0.085 | hhatla | -0.047 |