slingshot protein phosphatase 2a
ZFIN 
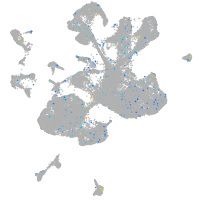
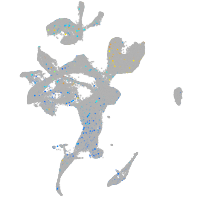
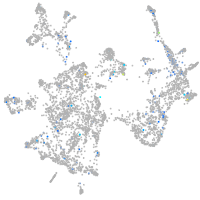
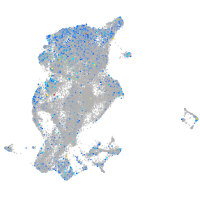
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| SPAG9 | 0.188 | uraha | -0.083 |
| si:dkeyp-82a1.2 | 0.176 | CABZ01021592.1 | -0.081 |
| kita | 0.142 | si:dkey-251i10.2 | -0.081 |
| itga3a | 0.136 | paics | -0.072 |
| tyr | 0.135 | mdh1aa | -0.063 |
| lamp1a | 0.132 | prdx5 | -0.060 |
| oca2 | 0.125 | phyhd1 | -0.058 |
| CU466278.1 | 0.125 | tmem130 | -0.054 |
| bace2 | 0.124 | aldob | -0.046 |
| slc29a3 | 0.123 | cyb5a | -0.045 |
| LOC108180743 | 0.123 | oacyl | -0.045 |
| cmklr1 | 0.122 | sult1st1 | -0.044 |
| gadd45bb | 0.121 | impdh1b | -0.042 |
| cers2a | 0.121 | krt18b | -0.041 |
| zgc:171490 | 0.121 | pvalb2 | -0.040 |
| yjefn3 | 0.120 | prdx1 | -0.039 |
| hgsnat | 0.120 | sesn1 | -0.038 |
| adssl1 | 0.120 | slc22a7a | -0.037 |
| dct | 0.119 | zgc:153031 | -0.037 |
| pmela | 0.119 | si:ch211-251b21.1 | -0.037 |
| pah | 0.119 | hbae3 | -0.036 |
| pim1 | 0.118 | hbbe1.3 | -0.035 |
| klf6a | 0.118 | pvalb1 | -0.035 |
| si:zfos-943e10.1 | 0.118 | slc2a15a | -0.034 |
| cited1 | 0.118 | ndrg2 | -0.033 |
| tyrp1a | 0.118 | gch2 | -0.033 |
| slc39a10 | 0.117 | gpd1b | -0.032 |
| atp6v0a2b | 0.117 | hbbe1.2 | -0.032 |
| mchr2 | 0.115 | aqp3a | -0.031 |
| osgn1 | 0.114 | TMEM19 | -0.031 |
| tyrp1b | 0.114 | calm1a | -0.030 |
| tfap2a | 0.114 | s100v2 | -0.030 |
| XLOC-009918 | 0.113 | hbae1.1 | -0.030 |
| tmem243b | 0.113 | iah1 | -0.030 |
| tfap2e | 0.112 | rbp4l | -0.030 |