secreted phosphoprotein 1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 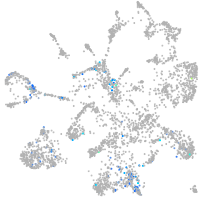 |
mesenchyme 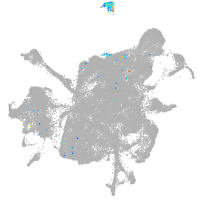 |
hematopoietic / vasculature 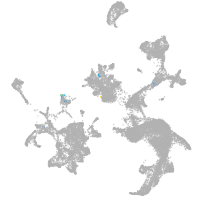 |
pronephros 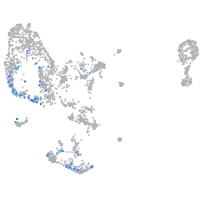 |
neural |
spinal cord / glia |
eye |
taste / olfactory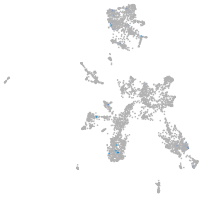 |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
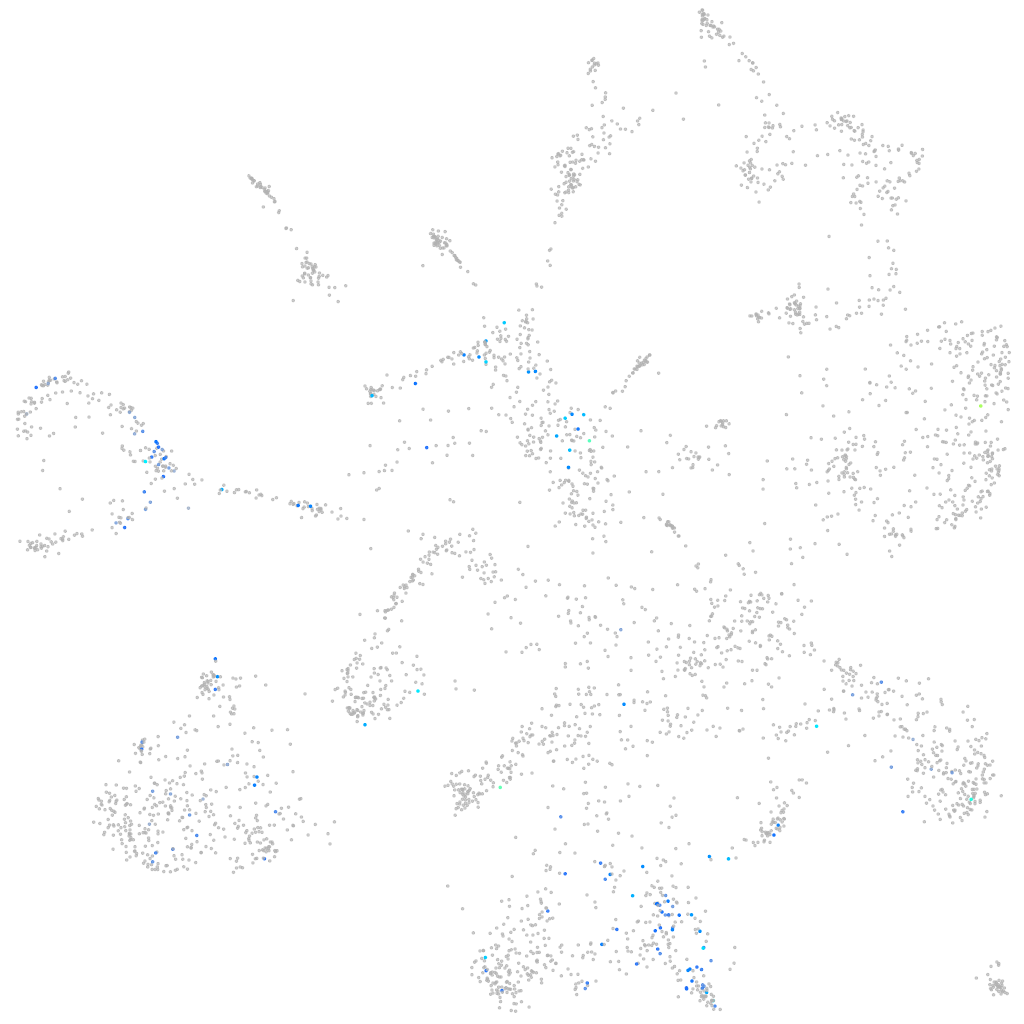
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kcnk18 | 0.219 | mdka | -0.133 |
| actc1a | 0.206 | krt94 | -0.101 |
| myh11a | 0.192 | tmem88b | -0.094 |
| BX649516.2 | 0.191 | jam2b | -0.088 |
| tpm1 | 0.186 | marcksl1b | -0.082 |
| tagln | 0.185 | tmem176 | -0.080 |
| acta2 | 0.184 | bambia | -0.078 |
| cald1b | 0.180 | rbp4 | -0.076 |
| acta1b | 0.177 | podxl | -0.072 |
| CR387919.1 | 0.176 | rpl36a | -0.071 |
| tuba8l2 | 0.175 | krt8 | -0.071 |
| acta1a | 0.175 | tgm2b | -0.071 |
| p2rx5 | 0.173 | fthl27 | -0.070 |
| mylkb | 0.172 | hspb1 | -0.070 |
| lmod1b | 0.171 | tmem98 | -0.069 |
| AL929457.1 | 0.168 | slc25a5 | -0.068 |
| csrp1b | 0.168 | krt15 | -0.068 |
| pln | 0.167 | ftr82 | -0.068 |
| tpm2 | 0.161 | cx43.4 | -0.064 |
| arr3b | 0.160 | pdgfra | -0.064 |
| hoxb1a | 0.158 | anxa1a | -0.063 |
| fbxl22 | 0.157 | cd151 | -0.063 |
| FO681390.1 | 0.156 | c1qtnf6a | -0.062 |
| tspan15 | 0.154 | ubc | -0.061 |
| LOC100331480 | 0.153 | lsp1 | -0.060 |
| pdlim3b | 0.153 | scarb2a | -0.059 |
| myl9a | 0.151 | bnc2 | -0.059 |
| atp2b1a | 0.151 | si:ch211-286o17.1 | -0.058 |
| cox6b1 | 0.148 | wu:fb55g09 | -0.058 |
| myl6 | 0.146 | si:ch211-39i22.1 | -0.058 |
| pnp4a | 0.146 | cebpd | -0.057 |
| mfsd4ab | 0.146 | gata5 | -0.057 |
| CR628365.1 | 0.145 | COLEC10 | -0.057 |
| kng1 | 0.145 | rps23 | -0.056 |
| casz1 | 0.144 | colec11 | -0.056 |