"spondin 2b, extracellular matrix protein"
ZFIN 
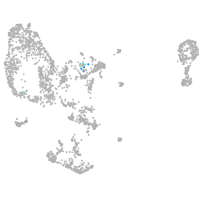
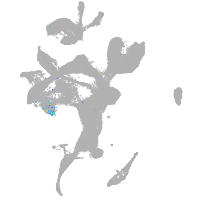
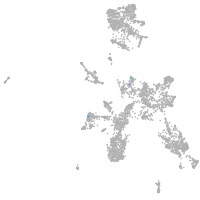
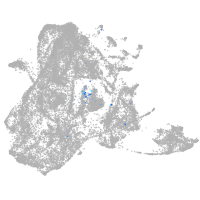
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| thbs4b | 0.281 | zgc:193541 | -0.037 |
| en1a | 0.264 | mdh1aa | -0.033 |
| CABZ01092746.1 | 0.263 | suclg1 | -0.033 |
| adamts17 | 0.244 | pgk1 | -0.033 |
| ecrg4a | 0.241 | pklr | -0.031 |
| ogna | 0.239 | gstr | -0.030 |
| fstb | 0.239 | scp2a | -0.030 |
| tbx18 | 0.209 | suclg2 | -0.029 |
| vwde | 0.196 | hadh | -0.029 |
| smpdl3a | 0.193 | slc37a4a | -0.029 |
| LOC100535130 | 0.184 | atp5f1b | -0.029 |
| c1qtnf5 | 0.184 | ckba | -0.028 |
| CABZ01041962.1 | 0.177 | mgst1.2 | -0.028 |
| ephx4 | 0.172 | dad1 | -0.028 |
| angptl1a | 0.170 | slc25a34 | -0.028 |
| tbx22 | 0.168 | rmdn1 | -0.028 |
| BX649411.2 | 0.167 | gpx4a | -0.028 |
| adgrf6 | 0.164 | hspe1 | -0.028 |
| prex2 | 0.164 | ndufb8 | -0.027 |
| twist1b | 0.162 | pgam1a | -0.027 |
| agtr2 | 0.162 | cox5ab | -0.027 |
| aldoca | 0.155 | echs1 | -0.026 |
| vstm4b | 0.154 | nipsnap3a | -0.026 |
| mir125a-1 | 0.151 | akr7a3 | -0.026 |
| tmem119b | 0.148 | dldh | -0.026 |
| scara3 | 0.145 | slc25a11 | -0.026 |
| isg15 | 0.145 | aldh7a1 | -0.026 |
| si:ch211-186j3.6 | 0.145 | cox6b1 | -0.026 |
| SCARF2 | 0.144 | gpd1b | -0.026 |
| tmem182a | 0.143 | atp5pb | -0.026 |
| si:ch211-196i2.1 | 0.143 | idh1 | -0.026 |
| XLOC-017534 | 0.143 | tdo2a | -0.026 |
| dcn | 0.143 | ephx5 | -0.025 |
| rerglb | 0.142 | acmsd | -0.025 |
| col1a1b | 0.140 | cyc1 | -0.025 |