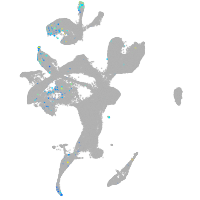
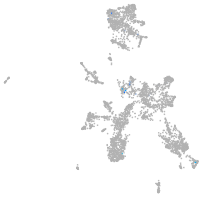
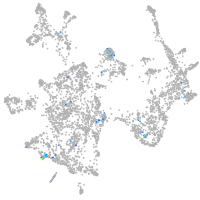
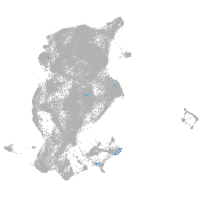
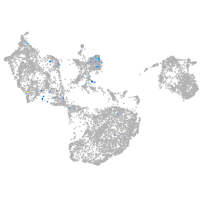
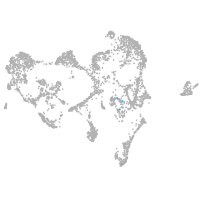
spondin 1a
ZFIN 
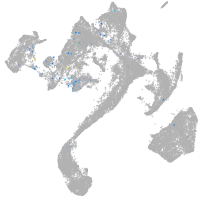
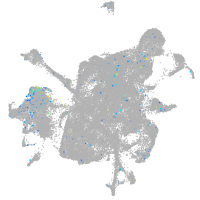
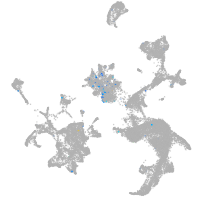
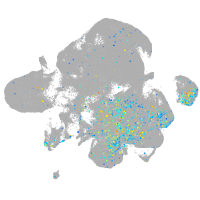
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gabrz | 0.348 | mat1a | -0.049 |
| gabra5 | 0.318 | si:dkey-16p21.8 | -0.045 |
| nyap2a | 0.306 | aqp12 | -0.045 |
| XLOC-025875 | 0.280 | apoc2 | -0.044 |
| XLOC-002533 | 0.270 | bhmt | -0.044 |
| ctsbb | 0.261 | mgst1.2 | -0.042 |
| cubn | 0.254 | gatm | -0.042 |
| fxyd6 | 0.253 | apoc1 | -0.041 |
| lrp2b | 0.252 | rpl30 | -0.040 |
| slitrk3b | 0.248 | rps13 | -0.040 |
| LOC100537369 | 0.243 | agxtb | -0.040 |
| tfeb | 0.243 | cdo1 | -0.039 |
| gfra2b | 0.242 | hspe1 | -0.039 |
| CABZ01087502.1 | 0.237 | ahcy | -0.039 |
| nlrp16 | 0.233 | rps15 | -0.039 |
| BX890548.1 | 0.232 | ybx1 | -0.039 |
| LOC100537272 | 0.231 | rps16 | -0.039 |
| ctsl.1 | 0.228 | rpl10a | -0.039 |
| XLOC-005257 | 0.224 | slc27a2a | -0.038 |
| amn | 0.214 | shmt1 | -0.037 |
| XLOC-036961 | 0.213 | glud1b | -0.037 |
| gabrd | 0.208 | gamt | -0.037 |
| lgmn | 0.205 | msrb2 | -0.037 |
| XLOC-040754 | 0.204 | grhprb | -0.036 |
| apba2a | 0.203 | serpinf2b | -0.036 |
| slc6a18 | 0.200 | rps26l | -0.036 |
| xirp1 | 0.198 | ambp | -0.036 |
| CABZ01079480.1 | 0.198 | fgb | -0.036 |
| TENM2 | 0.197 | igfbp2a | -0.036 |
| adam23a | 0.195 | fga | -0.036 |
| elmod1 | 0.195 | serpina1l | -0.036 |
| si:ch211-288d18.1 | 0.190 | ppdpfa | -0.036 |
| cacnb2a | 0.190 | ttc36 | -0.036 |
| XLOC-003375 | 0.187 | ces2 | -0.036 |
| mtbl | 0.186 | apobb.1 | -0.036 |