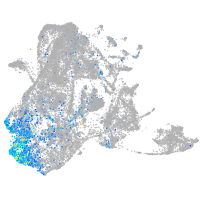
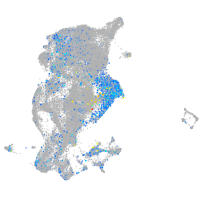
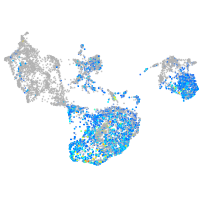
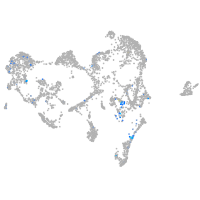
sp8 transcription factor b
ZFIN 
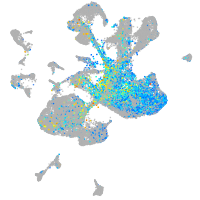
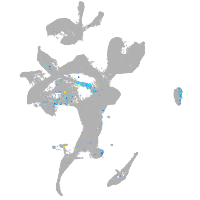
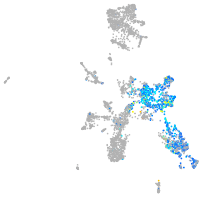
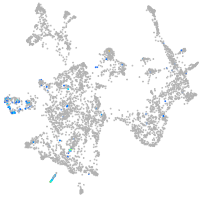
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sp8a | 0.356 | atp5mf | -0.055 |
| si:ch211-134a4.6 | 0.271 | atp5l | -0.053 |
| pcdh1g32 | 0.263 | mt-atp6 | -0.051 |
| LOC110438761 | 0.263 | mdh2 | -0.051 |
| CR848812.1 | 0.257 | mt-co2 | -0.049 |
| pax3a | 0.256 | BX000438.2 | -0.049 |
| nes | 0.253 | sod1 | -0.048 |
| her12 | 0.238 | cox8a | -0.047 |
| XLOC-020958 | 0.237 | COX3 | -0.046 |
| XLOC-039769 | 0.237 | atp5md | -0.046 |
| BX936393.1 | 0.237 | klf3 | -0.043 |
| BX323559.2 | 0.230 | mt-nd4 | -0.042 |
| neurog1 | 0.227 | atp5pf | -0.041 |
| prdm8 | 0.222 | cldnh | -0.041 |
| si:ch211-57n23.4 | 0.216 | ppifb | -0.040 |
| CR786582.1 | 0.208 | pvalb1 | -0.039 |
| dla | 0.206 | pvalb2 | -0.039 |
| her4.2 | 0.204 | atp6v1ba | -0.039 |
| mdka | 0.203 | foxp1b | -0.039 |
| XLOC-035300 | 0.203 | atp6v1h | -0.038 |
| XLOC-043447 | 0.202 | rnf128a | -0.038 |
| lpxn | 0.200 | b2ml | -0.038 |
| XLOC-003692 | 0.200 | echdc2 | -0.038 |
| prdm13 | 0.200 | gstp1 | -0.037 |
| filip1b | 0.198 | gsto2 | -0.037 |
| col14a1a | 0.197 | s100v2 | -0.037 |
| inavaa | 0.196 | rnaseka | -0.036 |
| mir219-3 | 0.194 | zgc:158463 | -0.035 |
| gsx1 | 0.191 | atp5f1e | -0.035 |
| lbx1b | 0.188 | itm2bb | -0.035 |
| BX530077.1 | 0.187 | cox7a2a | -0.035 |
| prtgb | 0.187 | LOC799574 | -0.035 |
| cpa6 | 0.187 | pitx2 | -0.035 |
| dlx5a | 0.186 | ca2 | -0.035 |
| dmbx1b | 0.184 | dedd1 | -0.034 |