SRY-box transcription factor 1b
ZFIN 
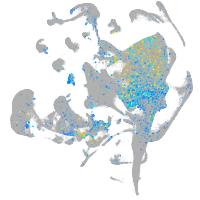
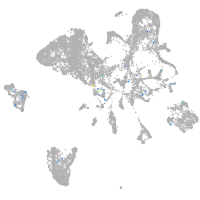
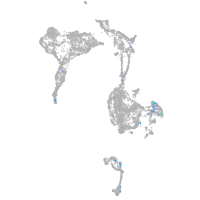
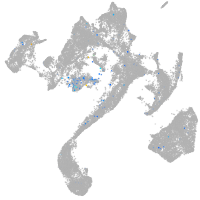
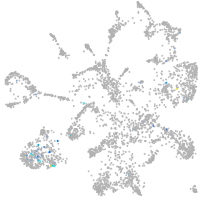
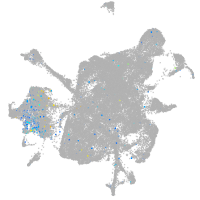
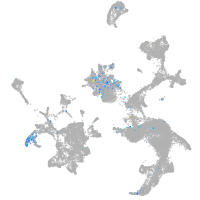
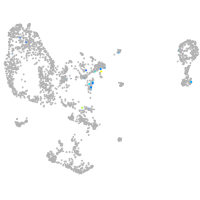
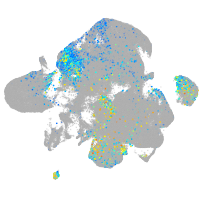
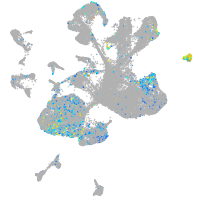
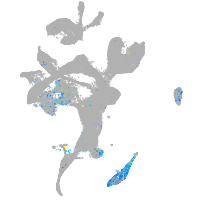
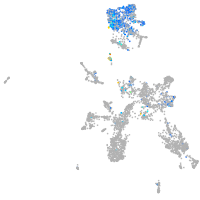
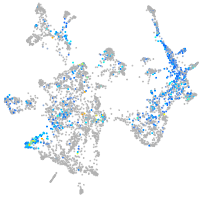
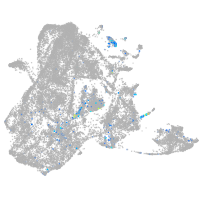
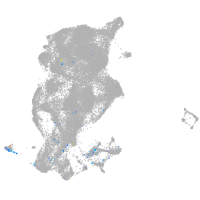
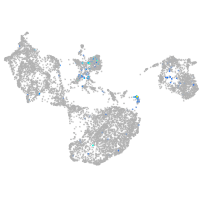
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sox1a | 0.389 | syngr1a | -0.069 |
| nkx2.4a | 0.314 | gpr143 | -0.060 |
| nkx2.4b | 0.263 | gmps | -0.059 |
| ptprna | 0.220 | smim29 | -0.059 |
| iqcg | 0.213 | paics | -0.059 |
| LOC103909971 | 0.205 | gstp1 | -0.057 |
| si:ch211-215j19.12 | 0.203 | glulb | -0.056 |
| ptprn2 | 0.200 | tspan36 | -0.055 |
| BX927275.2 | 0.197 | ctsd | -0.054 |
| LOC110438464 | 0.196 | krt18b | -0.052 |
| CABZ01079837.1 | 0.195 | cst14a.2 | -0.051 |
| CR855277.2 | 0.192 | zgc:153867 | -0.050 |
| si:ch73-139e5.2 | 0.192 | rnaseka | -0.050 |
| eno4 | 0.189 | LOC103910009 | -0.049 |
| si:dkey-222b8.4 | 0.188 | agtrap | -0.048 |
| scg2a | 0.187 | rab38 | -0.048 |
| ftr25 | 0.185 | actb2 | -0.048 |
| gdnfb | 0.184 | akr1b1 | -0.048 |
| scg2b | 0.181 | trpm1b | -0.047 |
| abhd8a | 0.179 | vamp3 | -0.047 |
| ano2 | 0.177 | prdx6 | -0.047 |
| htr1b | 0.175 | pttg1ipb | -0.047 |
| cnga2a | 0.173 | pfn1 | -0.047 |
| LOC100536937 | 0.169 | krt8 | -0.046 |
| CABZ01069436.1 | 0.166 | cyb5a | -0.045 |
| chd5 | 0.165 | bace2 | -0.045 |
| c1ql3b | 0.164 | gart | -0.045 |
| tor1l2 | 0.163 | BX571715.1 | -0.045 |
| ppp3cb | 0.161 | slc38a5b | -0.044 |
| rnasekb | 0.160 | prps1a | -0.044 |
| kif6 | 0.159 | ctsba | -0.044 |
| CABZ01015105.1 | 0.159 | dera | -0.044 |
| bsx | 0.158 | slc22a7a | -0.044 |
| gng3 | 0.158 | impdh1b | -0.043 |
| vat1l | 0.157 | bco1 | -0.043 |