SRY-box transcription factor 14
ZFIN 
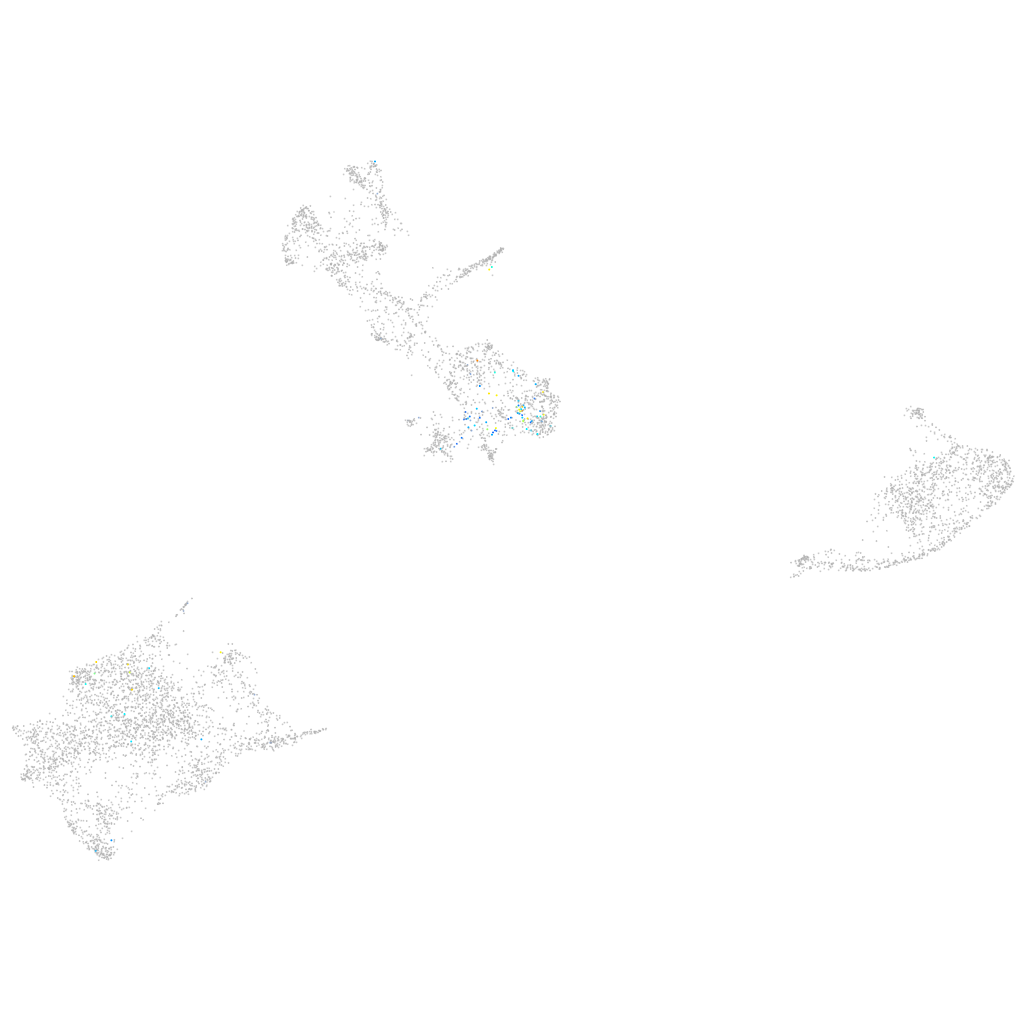
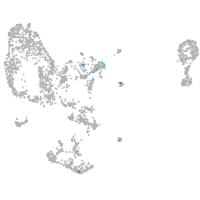
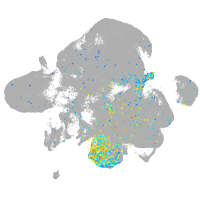
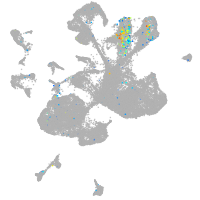
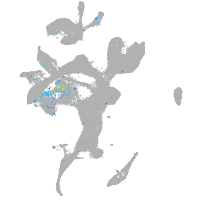
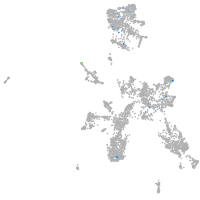
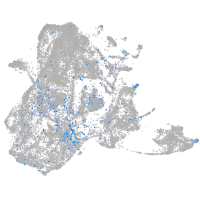
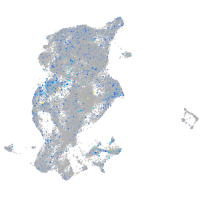
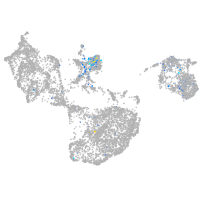
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tal2 | 0.271 | smim29 | -0.064 |
| zfpm2b | 0.247 | tspan36 | -0.064 |
| tal1 | 0.235 | syngr1a | -0.063 |
| grm1a | 0.234 | rnaseka | -0.060 |
| uncx | 0.231 | si:dkey-21a6.5 | -0.055 |
| lmo1 | 0.226 | gpr143 | -0.054 |
| otx2b | 0.205 | gmps | -0.053 |
| CR936482.1 | 0.198 | gstp1 | -0.053 |
| gpm6aa | 0.191 | pttg1ipb | -0.052 |
| slc32a1 | 0.181 | slc38a5b | -0.051 |
| gata3 | 0.181 | slc45a2 | -0.051 |
| dscamb | 0.180 | zgc:110239 | -0.050 |
| elavl3 | 0.177 | trpm1b | -0.049 |
| runx1t1 | 0.177 | ctsd | -0.049 |
| rbfox1 | 0.174 | ldhba | -0.048 |
| foxp2 | 0.171 | LOC103910009 | -0.048 |
| myt1la | 0.171 | rab32a | -0.048 |
| CABZ01067581.1 | 0.169 | eno3 | -0.047 |
| CABZ01076744.1 | 0.169 | prdx6 | -0.047 |
| nova2 | 0.167 | mitfa | -0.046 |
| zc4h2 | 0.167 | cdh1 | -0.046 |
| gpr85 | 0.167 | glulb | -0.046 |
| tuba1c | 0.165 | rab38 | -0.046 |
| slc6a1b | 0.164 | gyg1b | -0.046 |
| stmn1b | 0.164 | ctsba | -0.046 |
| inpp5d | 0.164 | cdk15 | -0.046 |
| gabrb4 | 0.163 | C12orf75 | -0.045 |
| lhx5 | 0.160 | opn5 | -0.045 |
| ZNF521 (1 of many) | 0.158 | degs1 | -0.044 |
| celf4 | 0.158 | cyth1b | -0.044 |
| lhx1a | 0.157 | atp11a | -0.043 |
| znf296 | 0.157 | si:zfos-943e10.1 | -0.043 |
| tenm3 | 0.157 | bace2 | -0.043 |
| FP236789.1 | 0.156 | rab34b | -0.043 |
| hcrtr2 | 0.156 | sypl2b | -0.042 |