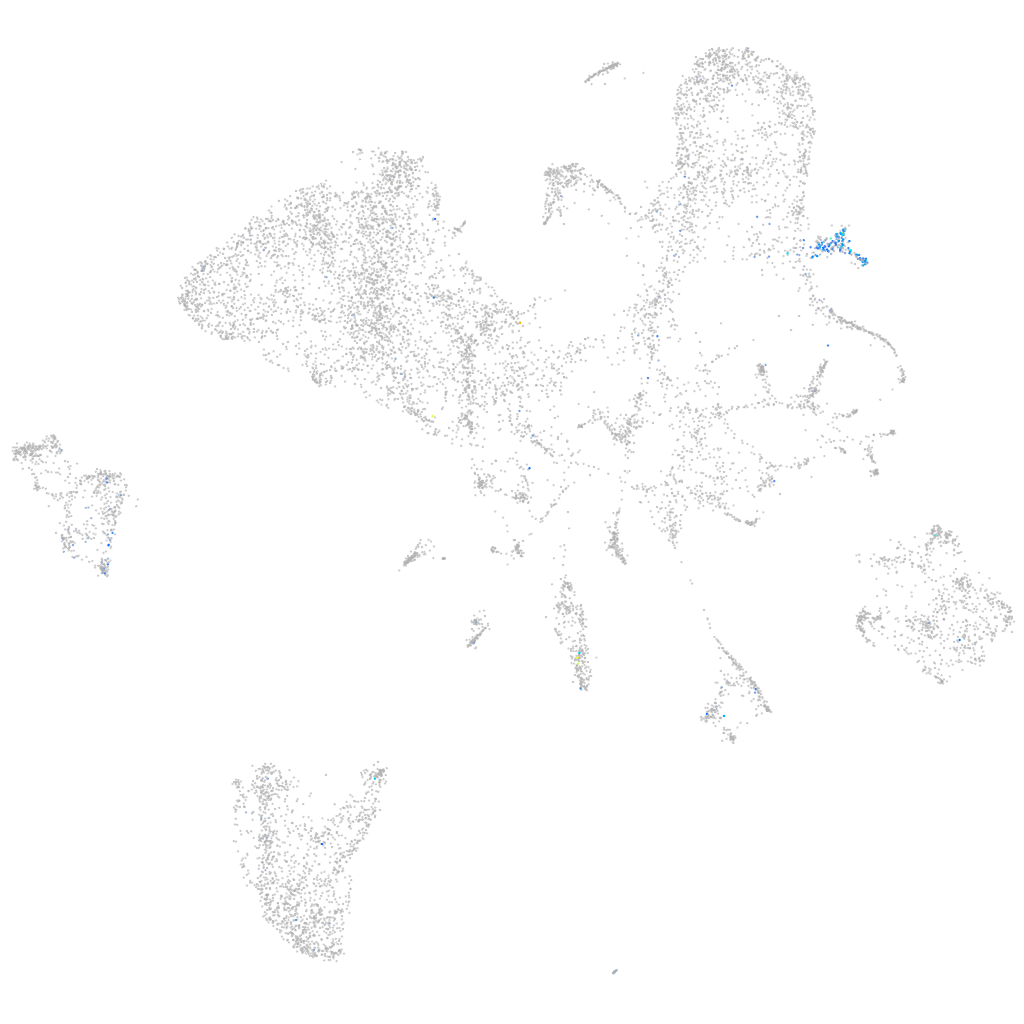
sorting nexin 8a
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cubn | 0.518 | mat1a | -0.078 |
| ctsbb | 0.510 | bhmt | -0.073 |
| lrp2b | 0.494 | aqp12 | -0.071 |
| LOC100537272 | 0.492 | apoa1b | -0.068 |
| dab2 | 0.440 | agxtb | -0.068 |
| amn | 0.434 | apoc2 | -0.067 |
| tfeb | 0.425 | kng1 | -0.066 |
| xirp1 | 0.423 | rbp2b | -0.065 |
| ctsl.1 | 0.405 | fgb | -0.065 |
| sptbn5 | 0.396 | ttc36 | -0.065 |
| lgmn | 0.394 | fga | -0.064 |
| slc6a18 | 0.382 | uox | -0.064 |
| ms4a17a.17 | 0.361 | pnp4b | -0.064 |
| scpep1 | 0.359 | serpinf2b | -0.064 |
| rab32b | 0.359 | hpda | -0.064 |
| naga | 0.353 | fabp10a | -0.063 |
| snx2 | 0.352 | apom | -0.063 |
| NAT16 (1 of many) | 0.350 | pygl | -0.062 |
| ctsh | 0.327 | grhprb | -0.062 |
| CU499336.2 | 0.327 | ttr | -0.062 |
| dpep1 | 0.322 | ambp | -0.062 |
| si:dkey-28n18.9 | 0.321 | gatm | -0.062 |
| epdl2 | 0.317 | vtnb | -0.062 |
| si:ch211-122f10.4 | 0.315 | apoc1 | -0.061 |
| tspan34 | 0.309 | apoa2 | -0.061 |
| cpvl | 0.304 | cpn1 | -0.061 |
| slc15a2 | 0.304 | cfb | -0.061 |
| acp7 | 0.296 | gc | -0.061 |
| tpte | 0.293 | fgg | -0.060 |
| CR381686.5 | 0.290 | si:ch73-281k2.5 | -0.060 |
| cdaa | 0.290 | cfhl4 | -0.060 |
| TMEM236 | 0.289 | ugp2a | -0.060 |
| fabp6 | 0.284 | ces2 | -0.060 |
| agtrap | 0.283 | serpinc1 | -0.060 |
| gnsb | 0.283 | shbg | -0.060 |