sorting nexin 6
ZFIN 
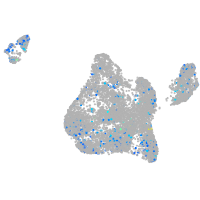
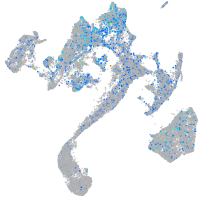
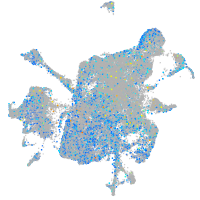
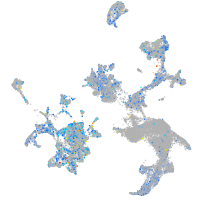
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp5fa1 | 0.178 | hspb1 | -0.141 |
| ppp1r15b-1 | 0.177 | wu:fb97g03 | -0.135 |
| mdh2 | 0.176 | apoeb | -0.125 |
| ldhba | 0.173 | ppig | -0.117 |
| atp1b1a | 0.171 | apoc1 | -0.115 |
| cox7a2a | 0.170 | cdx4 | -0.115 |
| mdh1aa | 0.168 | ncl | -0.115 |
| atp5mc1 | 0.168 | nucks1a | -0.114 |
| atp5meb | 0.165 | hnrnpabb | -0.113 |
| nnt | 0.164 | hmgb2a | -0.112 |
| eps8l3b | 0.164 | NC-002333.4 | -0.112 |
| sypl2a | 0.162 | rplp1 | -0.111 |
| dlst | 0.161 | akap12b | -0.110 |
| bin2a | 0.160 | gtf2f1 | -0.110 |
| GCA | 0.160 | npm1a | -0.108 |
| cpt1ab | 0.159 | zgc:110425 | -0.108 |
| got2b | 0.157 | hes6 | -0.108 |
| ppm1la | 0.156 | ebna1bp2 | -0.107 |
| pttg1ipb | 0.155 | parp1 | -0.106 |
| cdaa | 0.155 | rsl1d1 | -0.106 |
| glud1b | 0.155 | lmo2 | -0.106 |
| eno3 | 0.154 | zmp:0000000624 | -0.105 |
| mt-nd1 | 0.154 | hmgb2b | -0.105 |
| eif4ebp2 | 0.153 | si:ch73-1a9.3 | -0.104 |
| ndufa8 | 0.153 | si:ch73-281n10.2 | -0.103 |
| pdha1a | 0.152 | snu13b | -0.103 |
| cdh17 | 0.152 | gnl3 | -0.103 |
| atp5f1b | 0.152 | marcksb | -0.103 |
| stat5a | 0.151 | setb | -0.102 |
| cyc1 | 0.151 | nradd | -0.102 |
| lrrc66 | 0.151 | nop56 | -0.101 |
| mt-nd4 | 0.149 | msna | -0.101 |
| minos1 | 0.149 | rplp2l | -0.100 |
| BX323559.4 | 0.148 | hmga1a | -0.100 |
| cox6b1 | 0.147 | CT030188.1 | -0.099 |