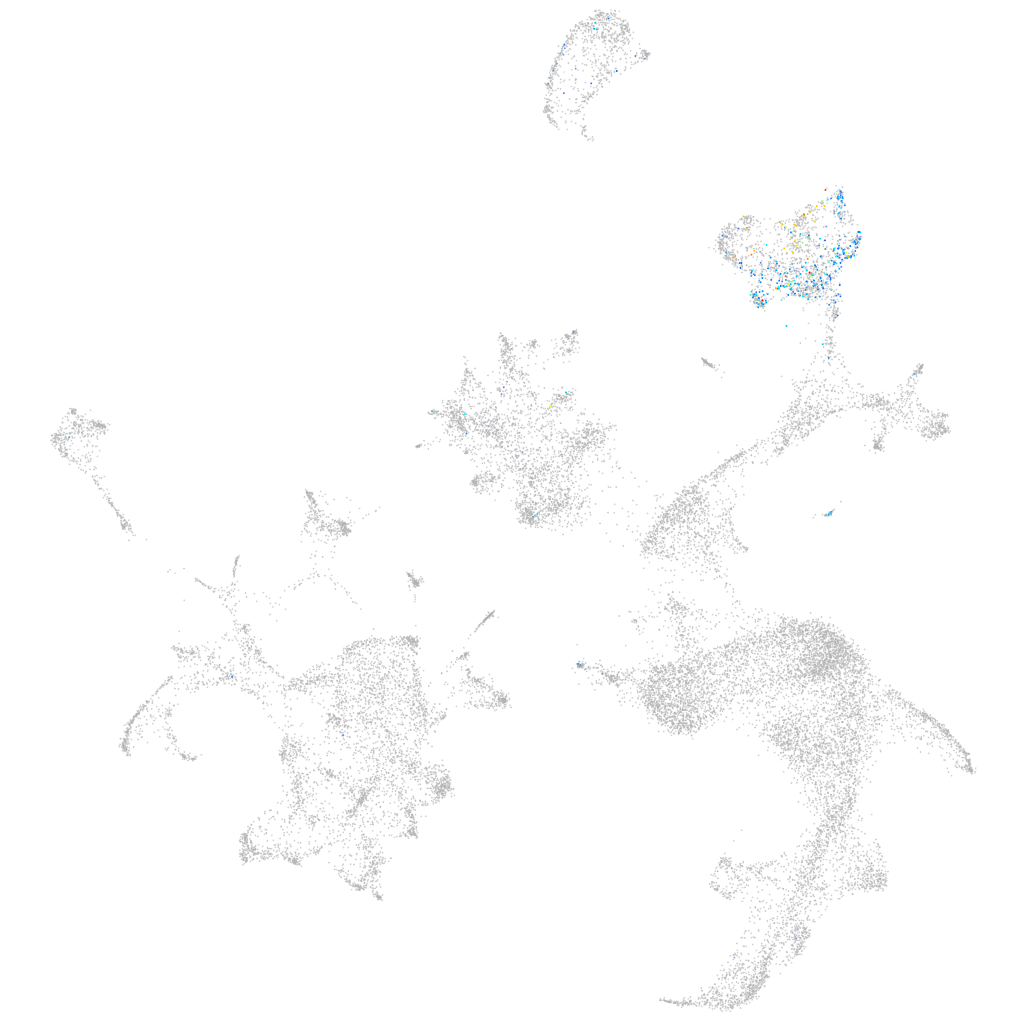
sorting nexin 10b
ZFIN 
Other cell groups


all cells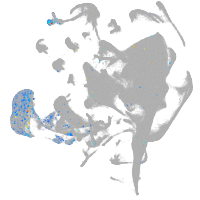 |
blastomeres |
PGCs |
endoderm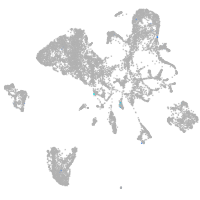 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 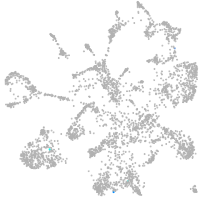 |
mesenchyme 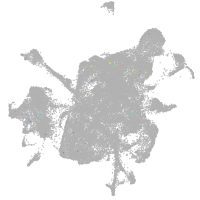 |
hematopoietic / vasculature 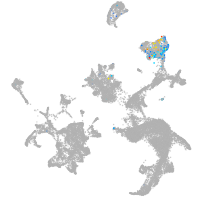 |
pronephros 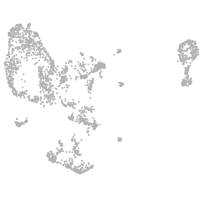 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line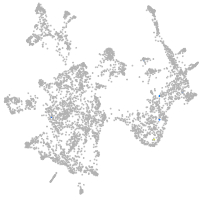 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-147m6.1 | 0.350 | hbae1.1 | -0.107 |
| si:ch211-194m7.3 | 0.345 | hbae3 | -0.102 |
| lygl1 | 0.342 | hbbe1.3 | -0.102 |
| ctss2.2 | 0.315 | hbae1.3 | -0.100 |
| lgals2a | 0.298 | hbbe2 | -0.100 |
| si:dkey-5n18.1 | 0.297 | hbbe1.1 | -0.095 |
| ctsl.1 | 0.296 | cahz | -0.090 |
| mfap4 | 0.282 | hbbe1.2 | -0.087 |
| acod1 | 0.274 | hemgn | -0.086 |
| stoml3b | 0.274 | zgc:56095 | -0.086 |
| ccl34a.4 | 0.268 | lmo2 | -0.084 |
| mpeg1.1 | 0.267 | si:ch211-250g4.3 | -0.081 |
| ms4a17a.10 | 0.267 | alas2 | -0.081 |
| tnfb | 0.266 | slc4a1a | -0.076 |
| cmklr1 | 0.265 | fth1a | -0.075 |
| card9 | 0.264 | creg1 | -0.075 |
| cx32.2 | 0.263 | nt5c2l1 | -0.075 |
| marco | 0.261 | zgc:163057 | -0.071 |
| cxcr3.2 | 0.258 | epb41b | -0.071 |
| grn1 | 0.258 | blvrb | -0.069 |
| slc2a6 | 0.257 | si:ch211-207c6.2 | -0.068 |
| si:ch211-153b23.7 | 0.254 | tspo | -0.067 |
| spi1a | 0.254 | aqp1a.1 | -0.066 |
| havcr1 | 0.252 | nmt1b | -0.065 |
| c1qb | 0.250 | plac8l1 | -0.062 |
| grna | 0.248 | tmod4 | -0.060 |
| gm2a | 0.243 | arl4aa | -0.059 |
| CR855311.1 | 0.242 | sptb | -0.058 |
| tspan36 | 0.242 | si:ch211-227m13.1 | -0.057 |
| cyba | 0.238 | znfl2a | -0.057 |
| c1qc | 0.234 | hbae5 | -0.057 |
| ctsba | 0.232 | uros | -0.056 |
| fcer1gl | 0.231 | tfr1a | -0.056 |
| si:zfos-1069f5.1 | 0.231 | hdr | -0.055 |
| cxcl19 | 0.230 | rfesd | -0.055 |