"SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1"
ZFIN 
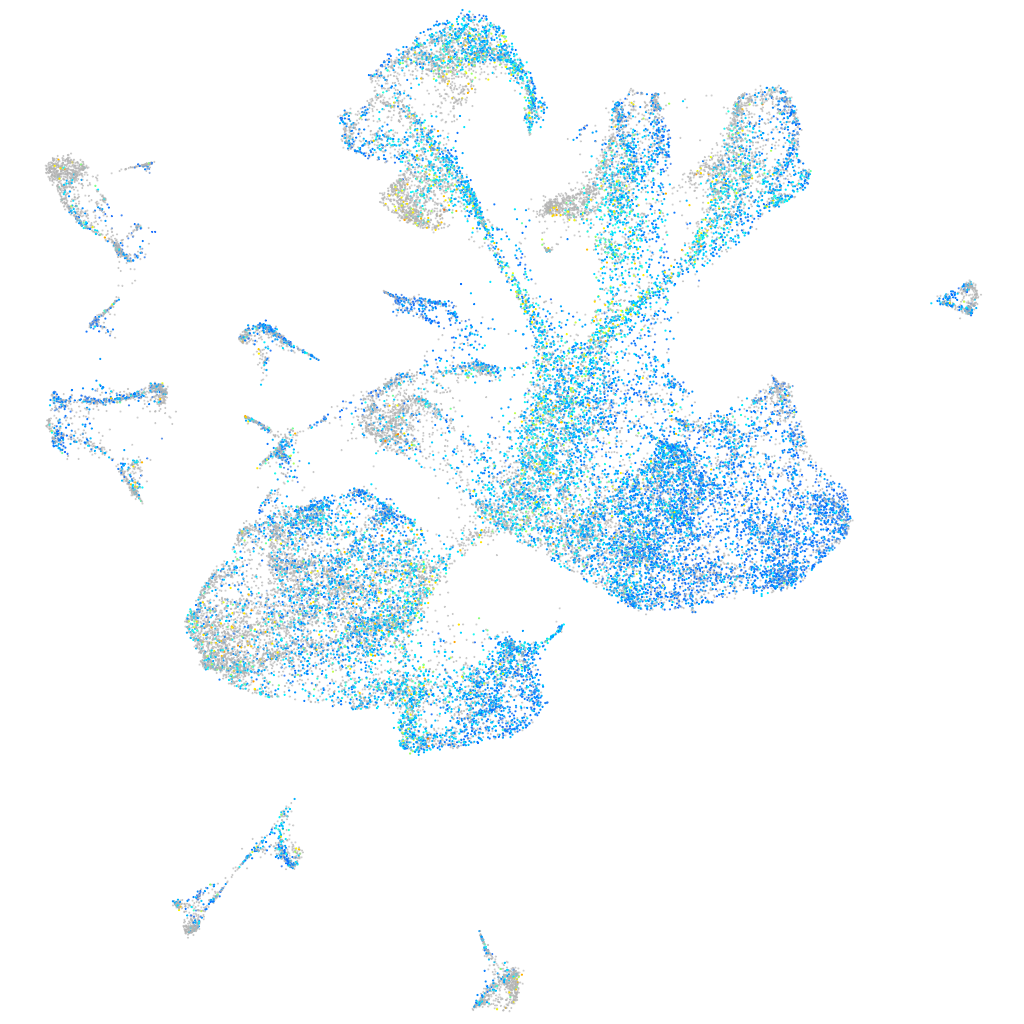
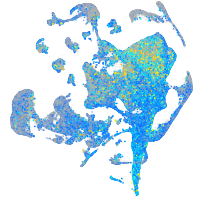
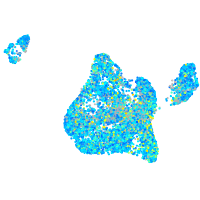
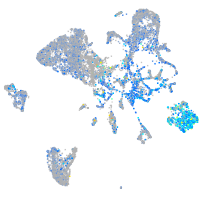
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpaba | 0.269 | glula | -0.137 |
| hnrnpa0l | 0.257 | atp1a1b | -0.137 |
| khdrbs1a | 0.248 | apoa2 | -0.129 |
| si:ch211-222l21.1 | 0.243 | cx43 | -0.124 |
| cirbpb | 0.241 | efhd1 | -0.121 |
| seta | 0.238 | apoa1b | -0.120 |
| ptmab | 0.231 | mt2 | -0.119 |
| hdac1 | 0.228 | cebpd | -0.118 |
| hmga1a | 0.227 | acbd7 | -0.118 |
| hnrnpa0b | 0.227 | slc1a2b | -0.115 |
| si:ch211-288g17.3 | 0.224 | si:ch211-66e2.5 | -0.112 |
| marcksb | 0.217 | slc4a4a | -0.111 |
| hnrnpabb | 0.215 | cox4i2 | -0.109 |
| eef2b | 0.215 | qki2 | -0.109 |
| cirbpa | 0.214 | ptn | -0.107 |
| sox11b | 0.214 | slc3a2a | -0.106 |
| syncrip | 0.208 | dap1b | -0.104 |
| tubb2b | 0.207 | ppap2d | -0.104 |
| hnrnpa0a | 0.206 | zgc:153704 | -0.103 |
| smarce1 | 0.204 | zwi | -0.101 |
| ddx39ab | 0.202 | mbpb | -0.101 |
| h3f3d | 0.202 | pvalb1 | -0.100 |
| nono | 0.201 | fabp2 | -0.100 |
| hspa8 | 0.199 | fabp7a | -0.100 |
| hmgn6 | 0.199 | prss59.2 | -0.099 |
| ilf2 | 0.198 | cldn7a | -0.099 |
| nucks1a | 0.198 | hepacama | -0.099 |
| chd4a | 0.197 | ptgdsb.2 | -0.098 |
| tpt1 | 0.193 | gapdhs | -0.098 |
| dlb | 0.192 | pvalb2 | -0.097 |
| hnrnpub | 0.192 | lpin1 | -0.097 |
| h2afvb | 0.192 | cdo1 | -0.096 |
| snrpd2 | 0.191 | mbpa | -0.096 |
| tp53inp2 | 0.191 | prss1 | -0.095 |
| eif3ea | 0.190 | ndrg3a | -0.094 |