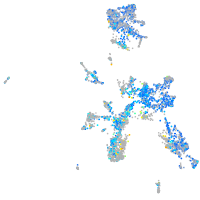
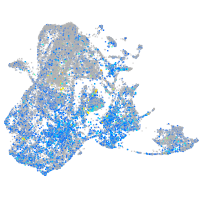
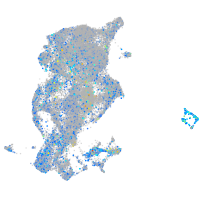
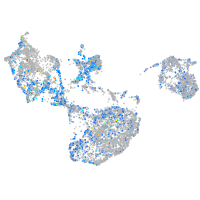
"SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1a"
ZFIN 
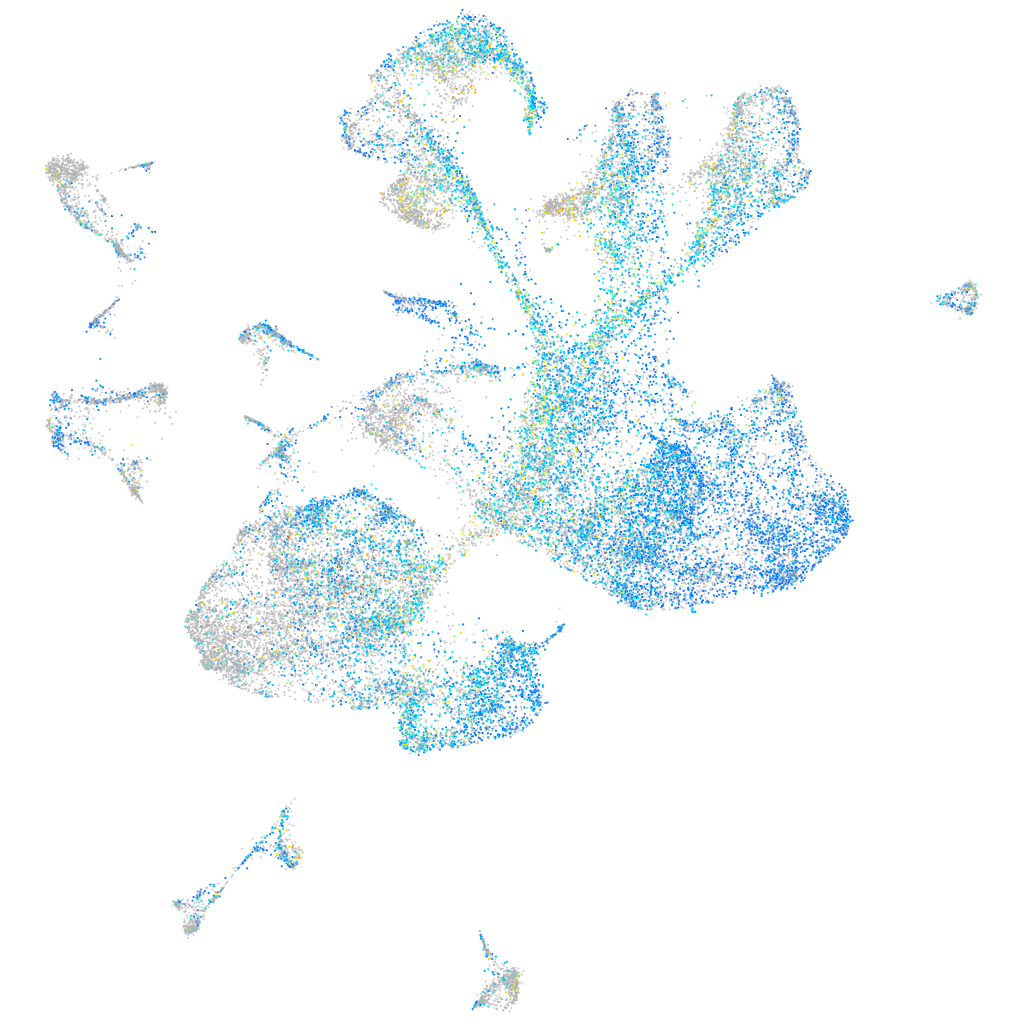
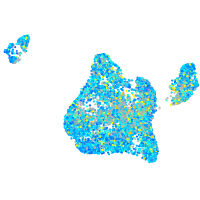
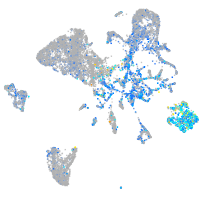
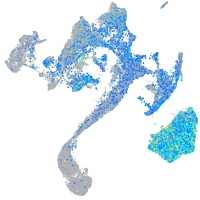
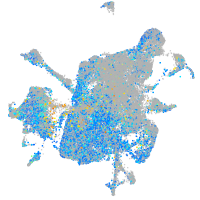
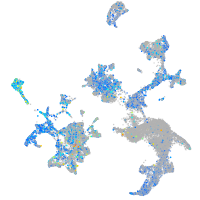
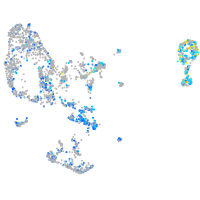
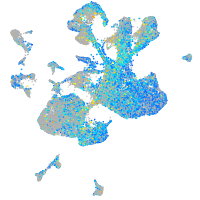
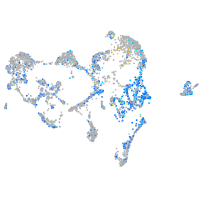
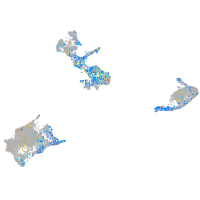
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-222l21.1 | 0.252 | glula | -0.166 |
| hnrnpaba | 0.248 | cx43 | -0.156 |
| ptmab | 0.245 | slc1a2b | -0.154 |
| hmga1a | 0.232 | si:ch211-66e2.5 | -0.150 |
| hnrnpa0l | 0.230 | efhd1 | -0.149 |
| khdrbs1a | 0.229 | atp1a1b | -0.149 |
| seta | 0.222 | ptn | -0.149 |
| cirbpb | 0.221 | ppap2d | -0.144 |
| cirbpa | 0.218 | acbd7 | -0.144 |
| marcksb | 0.217 | slc3a2a | -0.141 |
| si:ch211-288g17.3 | 0.215 | slc4a4a | -0.141 |
| sox11b | 0.207 | fabp7a | -0.139 |
| hdac1 | 0.206 | hepacama | -0.137 |
| h3f3a | 0.205 | cox4i2 | -0.136 |
| chd4a | 0.203 | ndrg3a | -0.134 |
| hnrnpa0b | 0.202 | cebpd | -0.133 |
| hnrnpabb | 0.201 | gpr37l1b | -0.132 |
| hmgb2b | 0.201 | anxa13 | -0.131 |
| tubb2b | 0.200 | gapdhs | -0.129 |
| h2afvb | 0.196 | smox | -0.129 |
| syncrip | 0.193 | qki2 | -0.128 |
| h3f3d | 0.193 | mt2 | -0.127 |
| ddx39ab | 0.190 | slc6a1b | -0.127 |
| nucks1a | 0.190 | cd63 | -0.126 |
| tmpob | 0.190 | apoa2 | -0.125 |
| nono | 0.189 | cdo1 | -0.124 |
| hmgn6 | 0.189 | sept8b | -0.121 |
| hmgn2 | 0.188 | ptgdsb.2 | -0.121 |
| dlb | 0.186 | zgc:153704 | -0.117 |
| smarca4a | 0.186 | slc1a3b | -0.117 |
| hsp90ab1 | 0.186 | aldocb | -0.114 |
| hnrnph1l | 0.183 | apoa1b | -0.114 |
| top1l | 0.182 | mgll | -0.113 |
| smarce1 | 0.181 | dap1b | -0.113 |
| ilf2 | 0.180 | cyp2ad3 | -0.113 |