slit homolog 3 (Drosophila)
ZFIN 
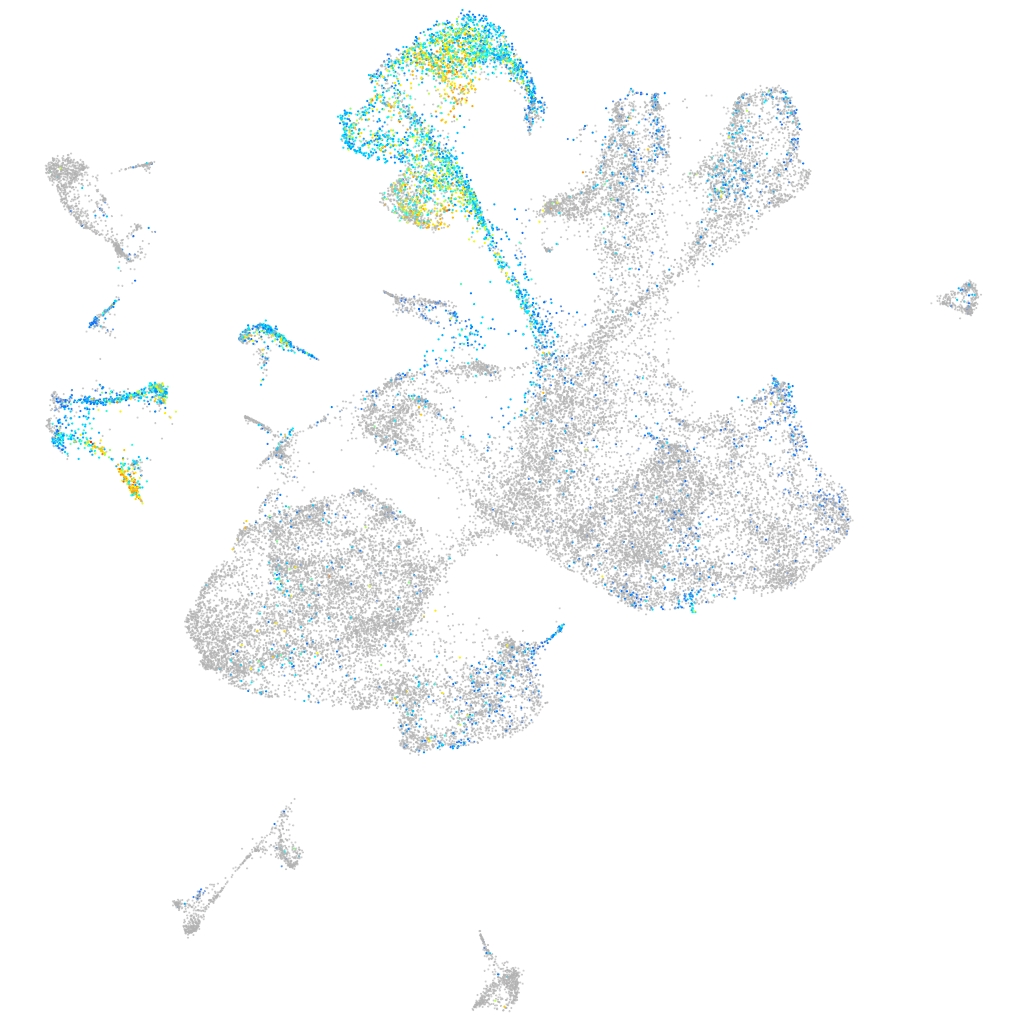
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc18a3a | 0.469 | XLOC-003690 | -0.283 |
| isl2a | 0.452 | sox3 | -0.274 |
| isl1 | 0.448 | XLOC-003689 | -0.250 |
| prph | 0.419 | her15.1 | -0.248 |
| chodl | 0.393 | id1 | -0.248 |
| islr2 | 0.391 | mdka | -0.240 |
| lhx4 | 0.367 | sox19a | -0.213 |
| tbx20 | 0.362 | sb:cb81 | -0.211 |
| phox2a | 0.342 | hmgb2a | -0.210 |
| chata | 0.341 | her4.2 | -0.206 |
| nrp1a | 0.338 | CU467822.1 | -0.201 |
| gap43 | 0.335 | XLOC-003692 | -0.199 |
| onecut1 | 0.333 | zgc:165461 | -0.198 |
| lmo4b | 0.330 | notch3 | -0.194 |
| slc5a7a | 0.330 | gas1b | -0.190 |
| phox2bb | 0.326 | lfng | -0.189 |
| tac1 | 0.313 | her4.1 | -0.183 |
| alcama | 0.308 | fabp7a | -0.180 |
| si:ch211-247j9.1 | 0.306 | notch1b | -0.179 |
| agrn | 0.303 | atp1a1b | -0.179 |
| elavl4 | 0.303 | gpm6bb | -0.176 |
| nefmb | 0.302 | dut | -0.175 |
| cntn2 | 0.300 | pcna | -0.175 |
| ache | 0.297 | si:ch73-21g5.7 | -0.173 |
| slit1b | 0.297 | cldn5a | -0.172 |
| map1b | 0.295 | zgc:56493 | -0.172 |
| vasna | 0.294 | msi1 | -0.172 |
| mnx1 | 0.291 | slc1a2b | -0.172 |
| xpr1a | 0.290 | abhd6a | -0.169 |
| sv2c | 0.289 | si:ch211-251b21.1 | -0.169 |
| rtn1b | 0.288 | btg2 | -0.168 |
| stmn2a | 0.282 | her12 | -0.167 |
| gng3 | 0.282 | her2 | -0.165 |
| chga | 0.281 | ranbp1 | -0.165 |
| clu | 0.277 | dla | -0.163 |