solute carrier family 43 member 2a
ZFIN 
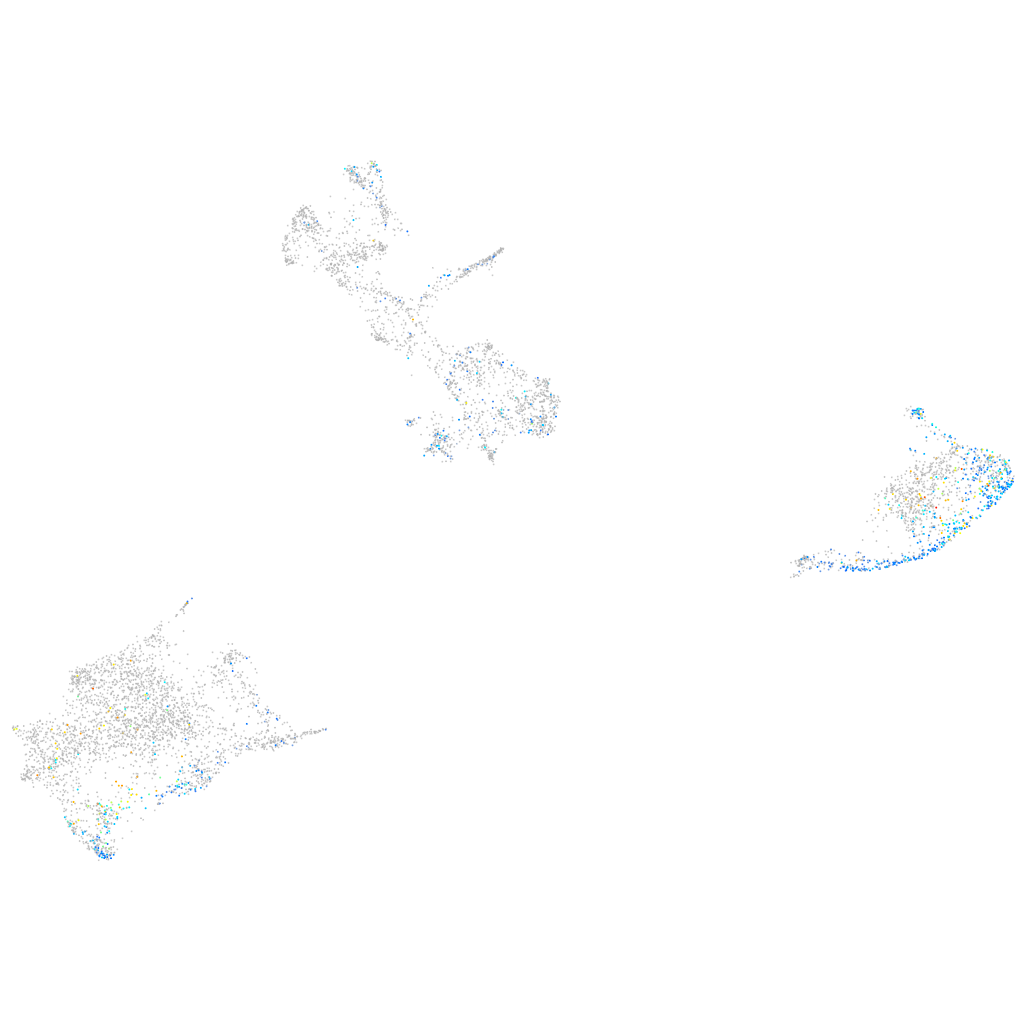
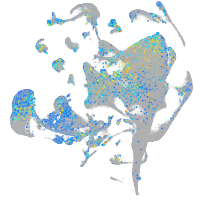
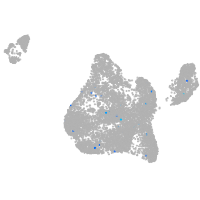
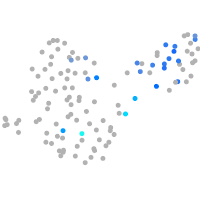
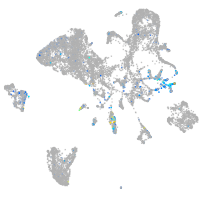
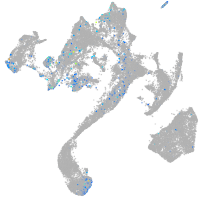
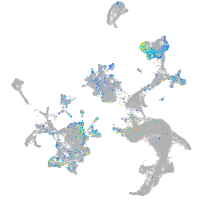
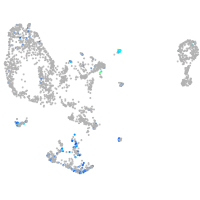
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kita | 0.321 | paics | -0.176 |
| SPAG9 | 0.303 | atic | -0.145 |
| oca2 | 0.299 | impdh1b | -0.140 |
| tyrp1a | 0.288 | uraha | -0.124 |
| slc24a5 | 0.287 | CABZ01021592.1 | -0.119 |
| pmela | 0.281 | defbl1 | -0.117 |
| tyr | 0.278 | prps1a | -0.115 |
| tyrp1b | 0.278 | mdh1aa | -0.114 |
| dct | 0.277 | apoda.1 | -0.114 |
| mlpha | 0.269 | prdx1 | -0.113 |
| kcnj13 | 0.261 | cyb5a | -0.111 |
| aadac | 0.259 | glulb | -0.104 |
| lamp1a | 0.258 | si:dkey-251i10.2 | -0.102 |
| slc29a3 | 0.255 | lypc | -0.102 |
| CDK18 | 0.253 | si:ch211-256m1.8 | -0.102 |
| slc22a2 | 0.245 | si:dkey-197i20.6 | -0.100 |
| opn5 | 0.243 | gpnmb | -0.100 |
| si:ch73-389b16.1 | 0.243 | hmgn2 | -0.094 |
| tfap2e | 0.243 | mdkb | -0.092 |
| zgc:91968 | 0.241 | tagln3b | -0.090 |
| megf10 | 0.239 | pltp | -0.090 |
| slc7a5 | 0.237 | unm-sa821 | -0.090 |
| mchr2 | 0.235 | si:ch73-89b15.3 | -0.089 |
| mtbl | 0.234 | ppat | -0.089 |
| slc24a4a | 0.233 | si:ch211-222l21.1 | -0.089 |
| cdh1 | 0.233 | rpl19 | -0.088 |
| slc39a10 | 0.230 | psat1 | -0.088 |
| CR847566.1 | 0.229 | shmt1 | -0.087 |
| atp6v0a2b | 0.225 | alx4a | -0.087 |
| rasd1 | 0.225 | gmps | -0.086 |
| slc3a2a | 0.222 | gapdhs | -0.085 |
| bace2 | 0.217 | adsl | -0.084 |
| si:ch211-195b13.1 | 0.216 | aqp1a.1 | -0.083 |
| tspan10 | 0.214 | guk1a | -0.083 |
| slc45a2 | 0.213 | s100v2 | -0.083 |