solute carrier family 43 member 2a
ZFIN 
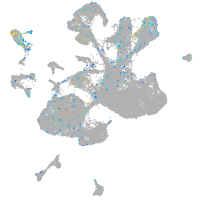
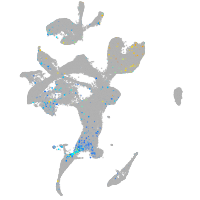
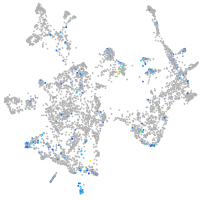
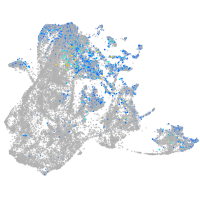
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sst2 | 0.388 | rps20 | -0.145 |
| zgc:195023 | 0.352 | rplp2l | -0.121 |
| nmba | 0.318 | gstr | -0.117 |
| kctd12.2 | 0.311 | cox6b1 | -0.114 |
| CU019662.1 | 0.288 | cx32.3 | -0.114 |
| sst1.2 | 0.281 | rps12 | -0.111 |
| pdyn | 0.267 | sod1 | -0.109 |
| insm1a | 0.267 | aldh7a1 | -0.107 |
| isl1 | 0.266 | si:dkey-151g10.6 | -0.106 |
| pax6b | 0.260 | ppa1b | -0.103 |
| scg3 | 0.257 | glud1b | -0.103 |
| gldn | 0.256 | rpl22l1 | -0.103 |
| dkk3b | 0.253 | nme2b.1 | -0.102 |
| si:dkey-153k10.9 | 0.252 | c1qbp | -0.102 |
| scg2b | 0.251 | sult2st2 | -0.101 |
| gpr4 | 0.248 | eno3 | -0.101 |
| pcsk1nl | 0.245 | hspe1 | -0.100 |
| LOC571123 | 0.241 | pa2g4a | -0.100 |
| scgn | 0.238 | npm1a | -0.100 |
| adgb | 0.236 | cox6a1 | -0.099 |
| napbb | 0.232 | ugt1a7 | -0.099 |
| camk1da | 0.229 | ahcy | -0.099 |
| kcnj11 | 0.229 | atp5mc3b | -0.099 |
| mir7a-1 | 0.227 | hspd1 | -0.098 |
| neurod1 | 0.227 | gstt1a | -0.098 |
| slc35g2b | 0.224 | rps3 | -0.097 |
| aldocb | 0.223 | gapdh | -0.097 |
| map3k15 | 0.222 | scp2a | -0.097 |
| CR774179.5 | 0.222 | rmdn1 | -0.097 |
| rprmb | 0.219 | nipsnap3a | -0.095 |
| vat1 | 0.217 | sod2 | -0.094 |
| scg2a | 0.216 | gcshb | -0.094 |
| gpc1a | 0.209 | aldob | -0.094 |
| tspan7b | 0.208 | cox8b | -0.093 |
| adm2a | 0.202 | aldh6a1 | -0.093 |