solute carrier family 38 member 4
ZFIN 
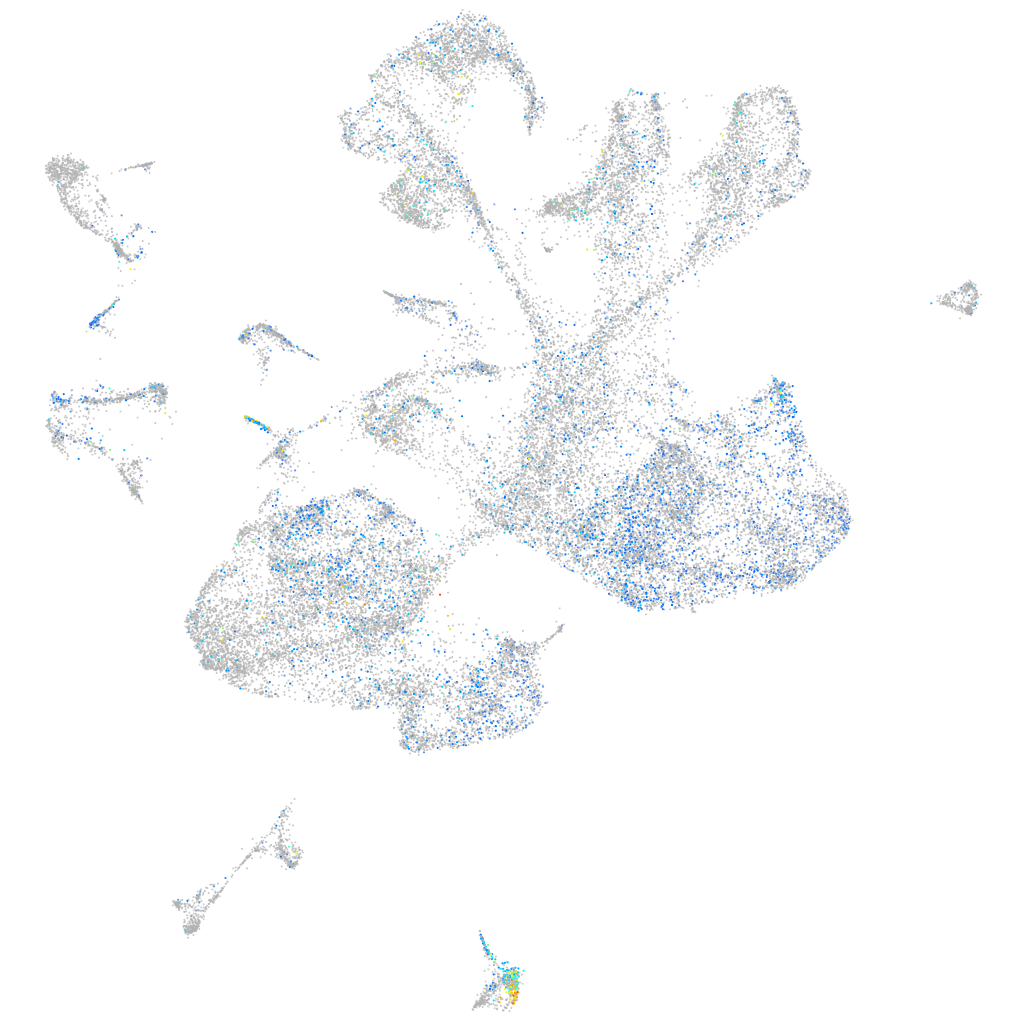
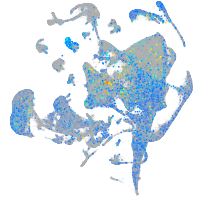
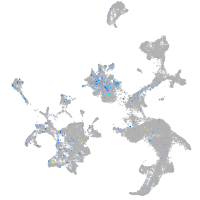
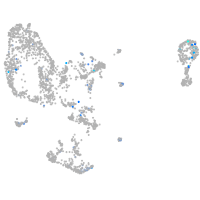
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ca14 | 0.238 | marcksl1b | -0.102 |
| slc2a1a | 0.232 | si:dkey-56m19.5 | -0.071 |
| col15a1b | 0.213 | elavl3 | -0.070 |
| vill | 0.198 | fabp3 | -0.066 |
| zgc:165604 | 0.195 | myt1b | -0.066 |
| rlbp1a | 0.193 | gpm6ab | -0.058 |
| hyal6 | 0.189 | tmsb4x | -0.053 |
| calml4b | 0.184 | stmn1b | -0.052 |
| fgf24 | 0.179 | atcaya | -0.051 |
| rbpms2b | 0.178 | myt1a | -0.051 |
| si:ch211-152c2.3 | 0.177 | gng2 | -0.051 |
| rhbg | 0.176 | zc4h2 | -0.050 |
| aqp4 | 0.169 | calm1a | -0.049 |
| cavin2a | 0.166 | si:dkey-276j7.1 | -0.049 |
| vil1 | 0.162 | syt11a | -0.049 |
| apoeb | 0.157 | slc6a1b | -0.048 |
| lmo7b | 0.149 | scrt2 | -0.048 |
| mgarp | 0.148 | nova2 | -0.048 |
| fzd5 | 0.148 | dpysl3 | -0.047 |
| rergla | 0.147 | rtn1a | -0.047 |
| gpr37a | 0.145 | cplx2 | -0.047 |
| eva1a | 0.144 | tuba8l3 | -0.046 |
| si:dkey-16p21.8 | 0.143 | csdc2a | -0.046 |
| tgfb3 | 0.142 | stxbp1a | -0.046 |
| rx3 | 0.136 | tubb5 | -0.045 |
| myo15aa | 0.135 | ubap2a | -0.045 |
| stc2a | 0.134 | gng3 | -0.045 |
| gyg1b | 0.134 | tuba1c | -0.045 |
| cahz | 0.133 | mllt11 | -0.045 |
| inhbab | 0.131 | jpt1b | -0.045 |
| CABZ01077555.1 | 0.129 | gdi1 | -0.045 |
| gstp1 | 0.127 | elavl4 | -0.045 |
| ush1c | 0.122 | tp53inp2 | -0.044 |
| espn | 0.122 | gabarapb | -0.044 |
| CU929418.2 | 0.121 | nhlh2 | -0.044 |