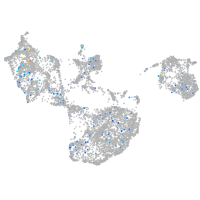
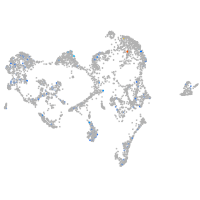
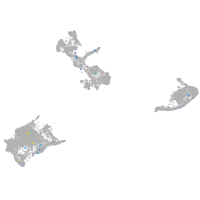
solute carrier family 37 member 4a
ZFIN 
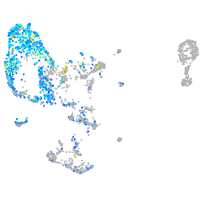
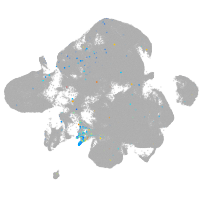
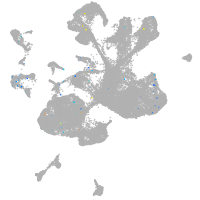
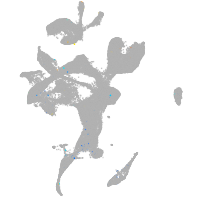
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU138547.1 | 0.716 | cd2bp2 | -0.254 |
| si:ch211-183d21.3 | 0.685 | acin1b | -0.253 |
| gstm.2 | 0.675 | ppig | -0.251 |
| trim25l | 0.638 | fbl | -0.240 |
| myofl | 0.630 | baz1b | -0.233 |
| LOC101884632 | 0.626 | snrpa | -0.232 |
| BX649423.1 | 0.624 | gtf2f1 | -0.228 |
| slit3 | 0.618 | h1m | -0.227 |
| CR847530.1 | 0.599 | hnrnpaba | -0.224 |
| rusc2 | 0.597 | ncl | -0.224 |
| enpp2 | 0.596 | cbx5 | -0.221 |
| CABZ01101984.1 | 0.593 | sde2 | -0.219 |
| cadps2 | 0.592 | tpx2 | -0.217 |
| ntn4 | 0.591 | tcea1 | -0.216 |
| agmo | 0.590 | u2surp | -0.216 |
| DPYSL2 (1 of many) | 0.590 | safb | -0.213 |
| prmt9 | 0.582 | snrpb2 | -0.208 |
| hs6st1a | 0.582 | dnttip2 | -0.207 |
| slc25a37 | 0.575 | hnrnpa1b | -0.206 |
| ppp1r9ba | 0.574 | si:dkeyp-69c1.6 | -0.204 |
| col2a1b | 0.562 | mphosph10 | -0.202 |
| LOC101883704 | 0.559 | hmga1a | -0.200 |
| dip2a | 0.557 | tbx16 | -0.199 |
| unc13d | 0.557 | NC-002333.4 | -0.198 |
| rcan3 | 0.557 | cactin | -0.191 |
| lrrc4ba | 0.548 | akap12b | -0.189 |
| smug1 | 0.548 | nop56 | -0.189 |
| crfb1 | 0.545 | etf1b | -0.187 |
| zmp:0000000662 | 0.540 | ctcf | -0.186 |
| kcnh1a | 0.540 | paf1 | -0.185 |
| glsl | 0.539 | odc1 | -0.184 |
| BX957317.2 | 0.538 | stm | -0.183 |
| zgc:64201 | 0.537 | nasp | -0.183 |
| prss23 | 0.533 | anp32b | -0.182 |
| dnah6 | 0.527 | marcksl1b | -0.181 |