solute carrier family 32 member 1
ZFIN 
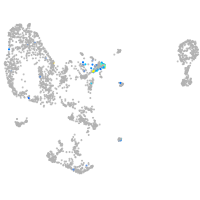
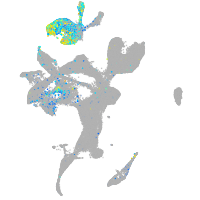
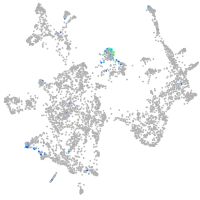
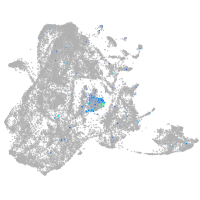
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| syt1a | 0.611 | hmgb2a | -0.352 |
| stxbp1a | 0.546 | pcna | -0.188 |
| slc6a1a | 0.530 | stmn1a | -0.187 |
| mir181b-3 | 0.525 | tuba8l4 | -0.185 |
| tfap2a | 0.503 | msna | -0.184 |
| stx1b | 0.500 | msi1 | -0.180 |
| sv2a | 0.489 | ahcy | -0.176 |
| slc6a1b | 0.449 | rrm1 | -0.173 |
| ywhag2 | 0.439 | dut | -0.173 |
| pax10 | 0.431 | mki67 | -0.171 |
| LOC101886114 | 0.431 | hmga1a | -0.170 |
| rbfox1 | 0.429 | nutf2l | -0.170 |
| elavl3 | 0.429 | ccnd1 | -0.170 |
| cplx3b | 0.429 | lbr | -0.168 |
| sncb | 0.428 | tuba8l | -0.167 |
| tfap2b | 0.417 | chaf1a | -0.165 |
| dtnbp1b | 0.410 | mcm7 | -0.162 |
| gad2 | 0.410 | selenoh | -0.161 |
| si:ch73-119p20.1 | 0.409 | ccna2 | -0.160 |
| gabrb4 | 0.407 | hmgb2b | -0.158 |
| cabp1b | 0.390 | banf1 | -0.156 |
| tiam1a | 0.389 | her15.1 | -0.155 |
| pvalb6 | 0.388 | fabp7a | -0.154 |
| celf5a | 0.377 | dek | -0.154 |
| eno2 | 0.367 | mdka | -0.152 |
| LOC100333482 | 0.361 | eef1da | -0.151 |
| nrxn1a | 0.359 | crx | -0.151 |
| gad1b | 0.359 | cks1b | -0.151 |
| stmn1b | 0.358 | otx5 | -0.148 |
| lrtm1 | 0.356 | rrm2 | -0.148 |
| vamp2 | 0.355 | rpa3 | -0.147 |
| pax6b | 0.353 | neurod4 | -0.145 |
| atp1b3a | 0.346 | lig1 | -0.145 |
| gng3 | 0.346 | fen1 | -0.145 |
| syn2a | 0.344 | mibp | -0.145 |