solute carrier family 30 member 2
ZFIN 
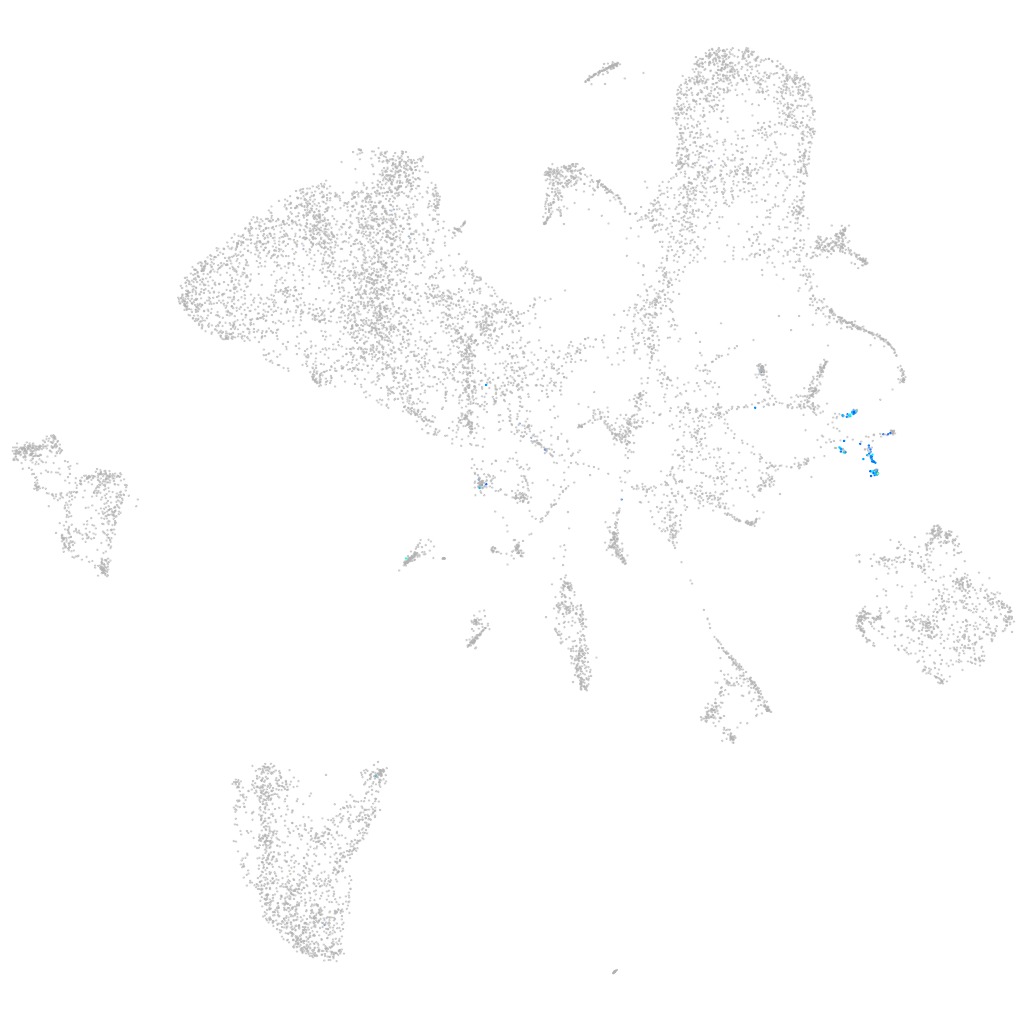
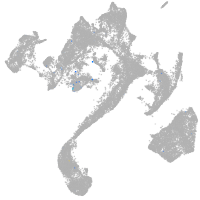
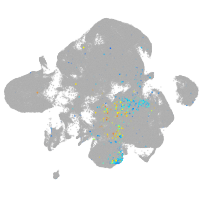
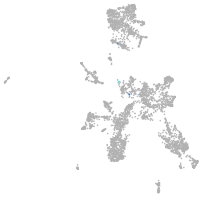
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ndufa4l2a | 0.652 | gapdh | -0.101 |
| SLC5A10 | 0.641 | ahcy | -0.099 |
| ptn | 0.604 | rps20 | -0.097 |
| ins | 0.603 | aldob | -0.095 |
| rims2a | 0.464 | gstp1 | -0.086 |
| scgn | 0.448 | gamt | -0.083 |
| pnoca | 0.430 | eno3 | -0.082 |
| gpr22a | 0.424 | gstr | -0.080 |
| cntnap2a | 0.424 | hdlbpa | -0.079 |
| pcsk2 | 0.406 | fbp1b | -0.077 |
| zgc:194990 | 0.395 | tuba8l2 | -0.077 |
| gcgb | 0.395 | nupr1b | -0.076 |
| pax6b | 0.394 | glud1b | -0.076 |
| pcsk1 | 0.393 | gatm | -0.075 |
| abhd15a | 0.386 | ckba | -0.073 |
| ucn3l | 0.381 | gstt1a | -0.072 |
| kcnj11 | 0.380 | gsta.1 | -0.072 |
| kcnj19a | 0.380 | si:dkey-16p21.8 | -0.072 |
| si:dkey-153k10.9 | 0.375 | lgals2b | -0.072 |
| plppr5b | 0.364 | aldh9a1a.1 | -0.071 |
| scg2a | 0.355 | fabp2 | -0.070 |
| dgkh | 0.355 | bhmt | -0.070 |
| pcsk1nl | 0.353 | dbi | -0.070 |
| uts1 | 0.352 | gpx4a | -0.070 |
| fam49bb | 0.351 | apoc2 | -0.070 |
| rprmb | 0.349 | apoa4b.1 | -0.069 |
| dkk3b | 0.346 | sod2 | -0.069 |
| gpr142 | 0.344 | sdr16c5b | -0.069 |
| CR774179.5 | 0.343 | aldh7a1 | -0.069 |
| neurod1 | 0.342 | apoa1b | -0.069 |
| npb | 0.341 | aldh6a1 | -0.068 |
| LOC100537384 | 0.338 | prdx6 | -0.068 |
| cpe | 0.336 | gnmt | -0.068 |
| disp2 | 0.335 | eef1da | -0.067 |
| cyp1c1 | 0.333 | cebpb | -0.067 |