solute carrier family 25 member 21
ZFIN 
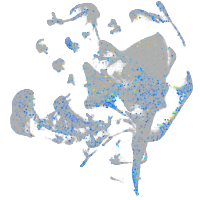
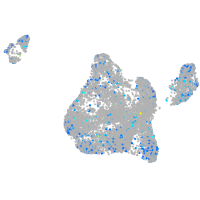
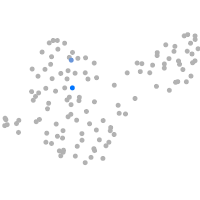
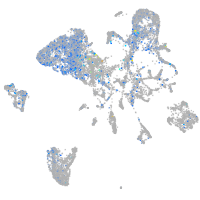
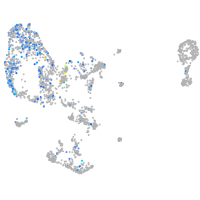
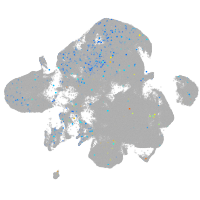
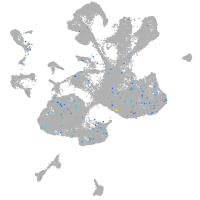
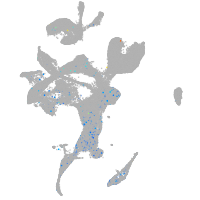
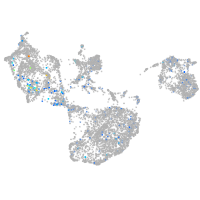
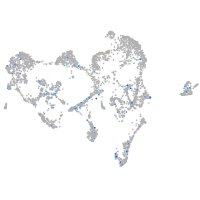
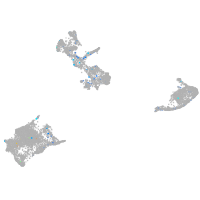
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| acmsd | 0.086 | elavl3 | -0.060 |
| grhprb | 0.085 | nova2 | -0.060 |
| agxta | 0.084 | tuba1c | -0.055 |
| ftcd | 0.084 | gpm6aa | -0.054 |
| hao1 | 0.084 | rtn1a | -0.053 |
| cx32.3 | 0.082 | tuba1a | -0.052 |
| dpys | 0.082 | stmn1b | -0.051 |
| fbp1b | 0.082 | gpm6ab | -0.049 |
| haao | 0.082 | mdkb | -0.049 |
| serpinf2b | 0.081 | ckbb | -0.046 |
| agxtb | 0.079 | myt1b | -0.044 |
| dpydb | 0.079 | tubb5 | -0.044 |
| f2 | 0.079 | gng3 | -0.043 |
| gapdh | 0.079 | hmgb3a | -0.042 |
| pbld | 0.079 | atp6v0cb | -0.041 |
| si:dkey-197j19.5 | 0.079 | myt1a | -0.041 |
| tymp | 0.079 | si:dkey-276j7.1 | -0.041 |
| aspdh | 0.078 | tmeff1b | -0.041 |
| eno3 | 0.078 | gng2 | -0.040 |
| hao2 | 0.078 | rnasekb | -0.040 |
| pklr | 0.078 | scrt2 | -0.040 |
| g6pca.2 | 0.077 | rtn1b | -0.039 |
| kng1 | 0.077 | sncb | -0.039 |
| nipsnap3a | 0.077 | dpysl3 | -0.038 |
| ahcy | 0.076 | epb41a | -0.038 |
| gstr | 0.075 | zc4h2 | -0.038 |
| serpinf2a | 0.075 | fez1 | -0.037 |
| slc37a4a | 0.075 | tmsb | -0.037 |
| urad | 0.075 | cspg5a | -0.036 |
| cyp4v8 | 0.074 | elavl4 | -0.036 |
| hal | 0.074 | vamp2 | -0.036 |
| rdh1 | 0.074 | dpysl2b | -0.036 |
| si:ch211-284e20.8 | 0.074 | nsg2 | -0.034 |
| slco1d1 | 0.074 | stx1b | -0.034 |
| upb1 | 0.074 | celf3a | -0.033 |