solute carrier family 24 member 5
ZFIN 
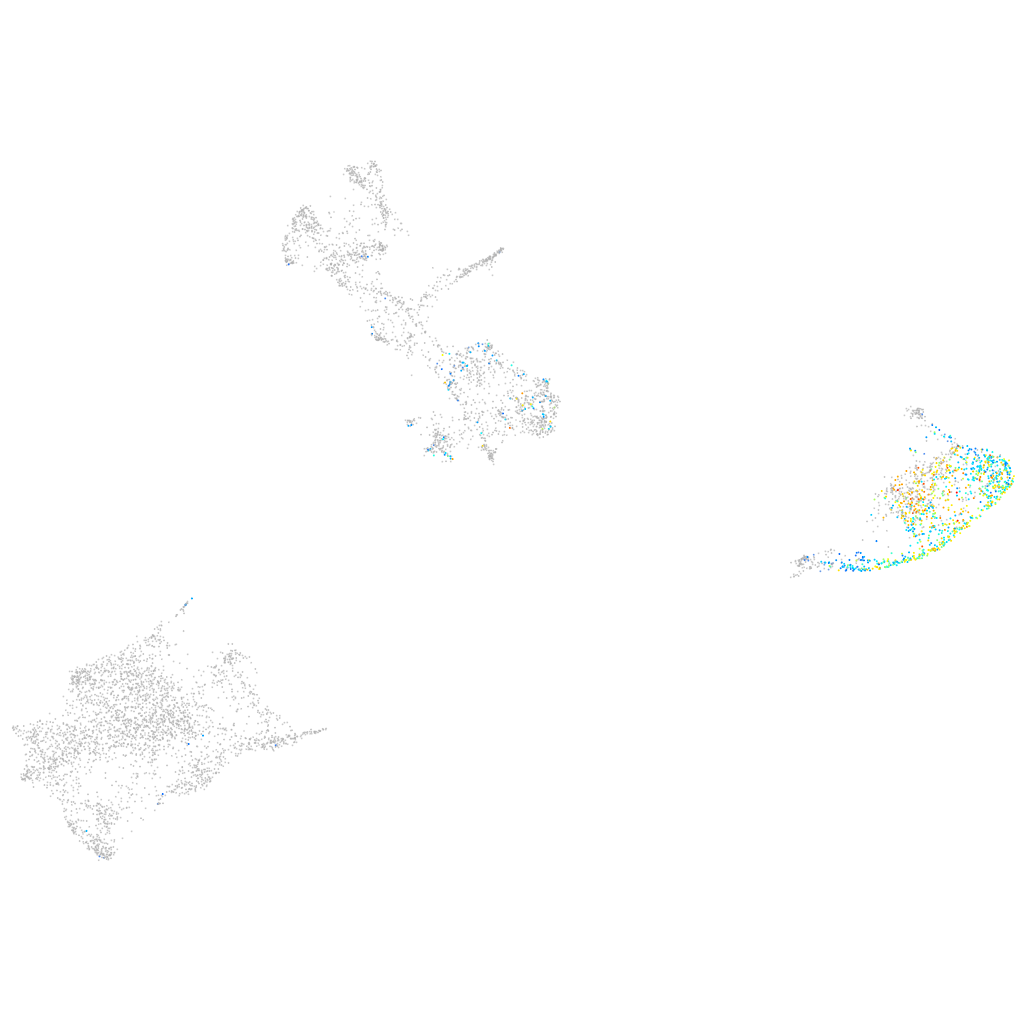
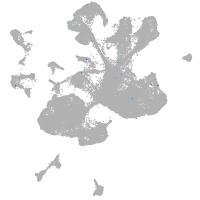
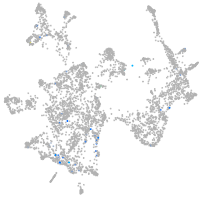
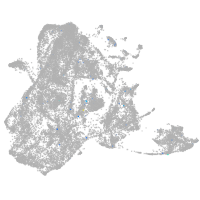
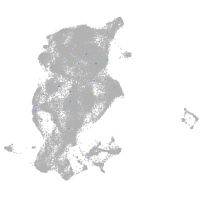
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tyrp1b | 0.753 | paics | -0.405 |
| pmela | 0.745 | CABZ01021592.1 | -0.395 |
| dct | 0.745 | uraha | -0.345 |
| tyrp1a | 0.741 | si:dkey-251i10.2 | -0.310 |
| zgc:91968 | 0.701 | mdh1aa | -0.302 |
| oca2 | 0.700 | tmem130 | -0.291 |
| kita | 0.631 | slc2a15a | -0.281 |
| mtbl | 0.616 | impdh1b | -0.277 |
| tyr | 0.599 | atic | -0.270 |
| si:ch73-389b16.1 | 0.597 | glulb | -0.266 |
| slc45a2 | 0.591 | phyhd1 | -0.261 |
| prkar1b | 0.583 | aox5 | -0.253 |
| aadac | 0.567 | si:ch211-251b21.1 | -0.246 |
| anxa1a | 0.538 | prps1a | -0.234 |
| slc22a2 | 0.533 | bscl2l | -0.222 |
| gstt1a | 0.530 | cyb5a | -0.221 |
| slc39a10 | 0.528 | pax7b | -0.213 |
| slc7a5 | 0.507 | pax7a | -0.206 |
| lamp1a | 0.497 | ppat | -0.202 |
| SPAG9 | 0.493 | sprb | -0.200 |
| kcnj13 | 0.492 | gmps | -0.199 |
| slc29a3 | 0.491 | krt18b | -0.198 |
| tspan10 | 0.487 | bco1 | -0.193 |
| tspan36 | 0.476 | shmt1 | -0.192 |
| slc37a2 | 0.472 | cx30.3 | -0.192 |
| mlpha | 0.462 | sult1st1 | -0.192 |
| slc3a2a | 0.457 | aldob | -0.188 |
| atp6v0ca | 0.456 | TMEM19 | -0.185 |
| atp6v0a2b | 0.449 | cax1 | -0.181 |
| hsd20b2 | 0.447 | defbl1 | -0.180 |
| slc24a4a | 0.443 | mibp | -0.178 |
| CDK18 | 0.443 | prdx1 | -0.177 |
| zdhhc2 | 0.442 | apoda.1 | -0.177 |
| AL935199.1 | 0.437 | prdx5 | -0.173 |
| pah | 0.435 | si:ch211-194k22.8 | -0.172 |