stem-loop binding protein
ZFIN 
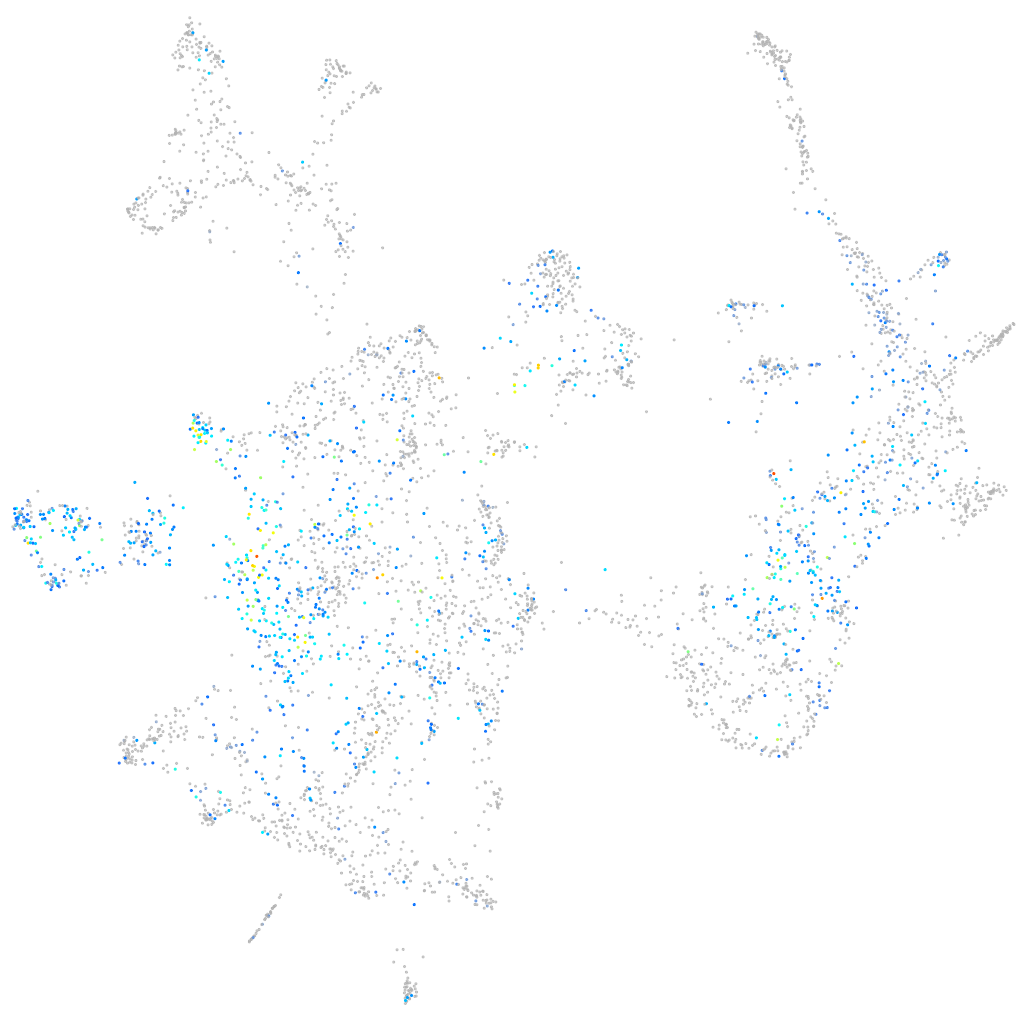
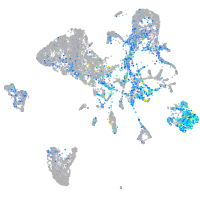
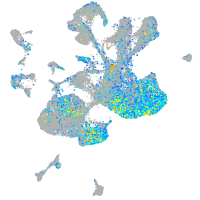
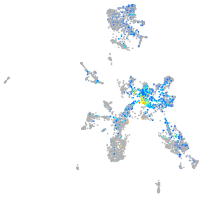
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.654 | h1f0 | -0.220 |
| rrm2 | 0.591 | tob1b | -0.210 |
| fen1 | 0.584 | atp2b1a | -0.204 |
| rpa2 | 0.572 | tmem59 | -0.187 |
| rrm1 | 0.569 | COX3 | -0.185 |
| nasp | 0.567 | b2ml | -0.182 |
| CABZ01005379.1 | 0.563 | rnasekb | -0.181 |
| dut | 0.559 | nupr1a | -0.179 |
| chaf1a | 0.554 | gapdhs | -0.179 |
| zgc:110540 | 0.553 | mt-co2 | -0.178 |
| cdc6 | 0.534 | calm1a | -0.177 |
| asf1ba | 0.530 | otofb | -0.173 |
| stmn1a | 0.520 | pvalb1 | -0.170 |
| mcm7 | 0.513 | calm1b | -0.169 |
| rpa3 | 0.510 | ccni | -0.166 |
| lig1 | 0.510 | gpx2 | -0.166 |
| unga | 0.505 | cd164l2 | -0.166 |
| dnajc9 | 0.501 | calml4a | -0.165 |
| orc4 | 0.498 | nptna | -0.163 |
| dhfr | 0.498 | dnajc5b | -0.162 |
| rfc4 | 0.475 | atp1a3b | -0.162 |
| gins2 | 0.475 | eno1a | -0.162 |
| esco2 | 0.474 | pvalb8 | -0.161 |
| shmt1 | 0.470 | kif1aa | -0.161 |
| LOC100330864 | 0.470 | osbpl1a | -0.159 |
| tk1 | 0.468 | CR383676.1 | -0.159 |
| sumo3b | 0.468 | pvalb2 | -0.159 |
| mcm4 | 0.463 | CR925719.1 | -0.157 |
| mcm5 | 0.463 | ip6k2a | -0.156 |
| mibp | 0.459 | pcsk5a | -0.156 |
| tubb2b | 0.455 | gabarapl2 | -0.155 |
| mcm2 | 0.454 | tspan13b | -0.155 |
| suv39h1b | 0.450 | atp6v1e1b | -0.154 |
| mcm3 | 0.450 | lrrc73 | -0.153 |
| rpa1 | 0.449 | apba1a | -0.153 |