SIX homeobox 2a
ZFIN 
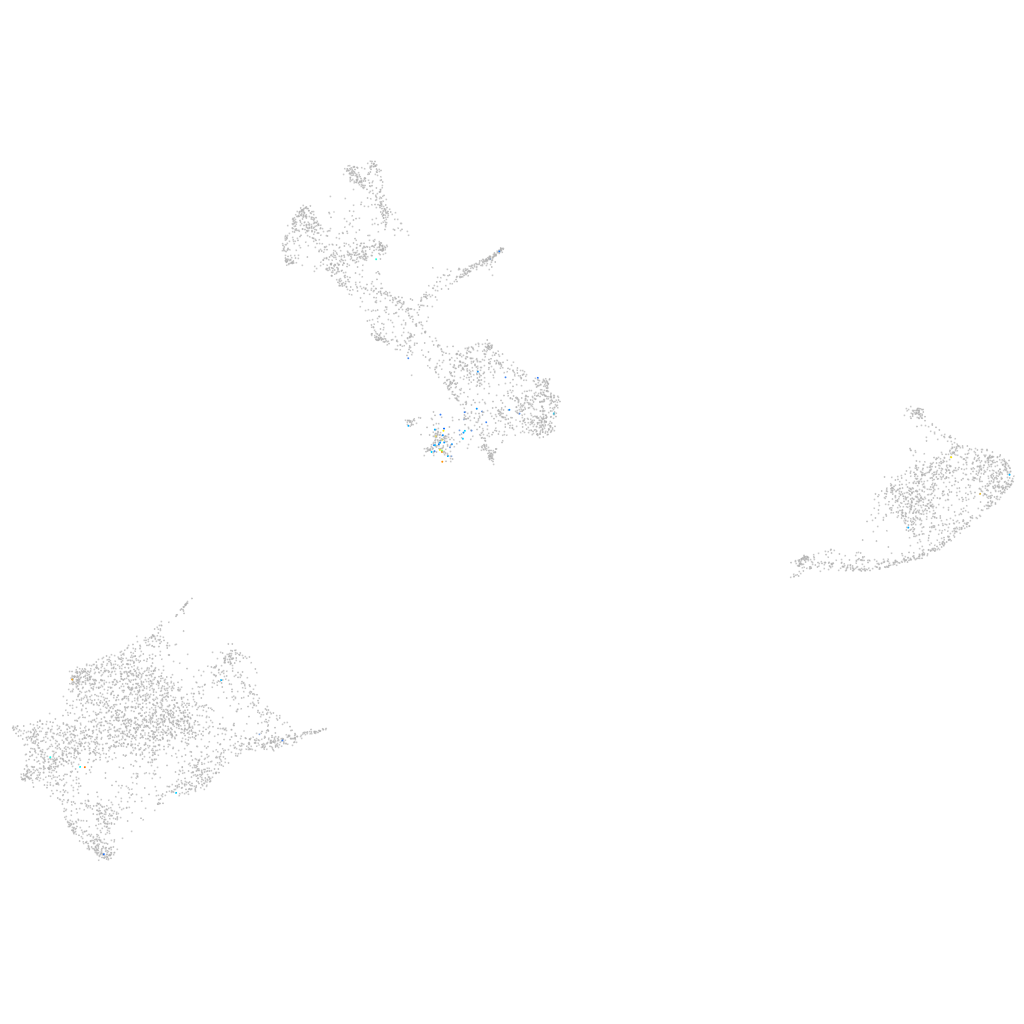
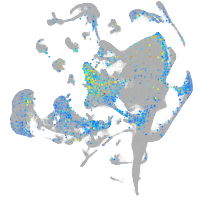
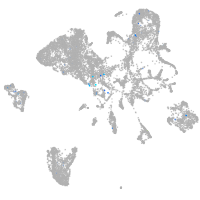
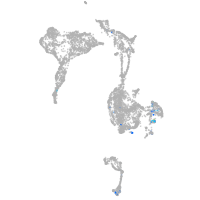
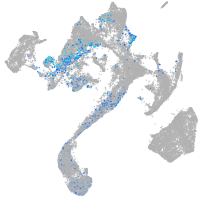
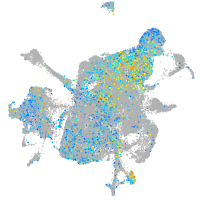
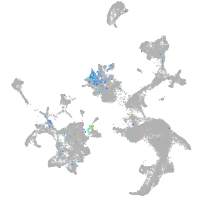
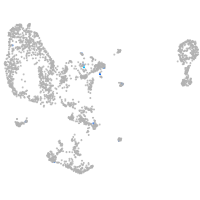
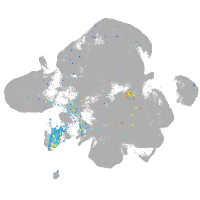
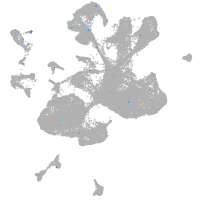
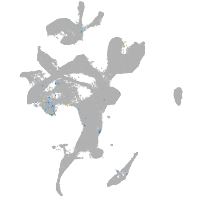
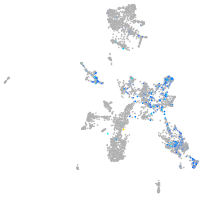
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc13a4 | 0.277 | syngr1a | -0.046 |
| ugt1b1 | 0.269 | tspan36 | -0.045 |
| foxc1b | 0.257 | impdh1b | -0.044 |
| XLOC-014486 | 0.252 | prps1a | -0.042 |
| fibina | 0.248 | akr1b1 | -0.042 |
| sim2 | 0.233 | agtrap | -0.036 |
| snx10b | 0.232 | rab32a | -0.036 |
| pde11al | 0.225 | atp11a | -0.036 |
| or137-4 | 0.224 | gmps | -0.034 |
| hspa12b | 0.219 | gpr143 | -0.033 |
| tmc8 | 0.219 | paics | -0.033 |
| si:ch73-24k9.2 | 0.212 | gpd1b | -0.032 |
| si:ch211-120e1.7 | 0.212 | atic | -0.032 |
| coch | 0.211 | prdx1 | -0.031 |
| tgfbi | 0.209 | tpd52 | -0.031 |
| LOC564253 | 0.207 | pcbd1 | -0.031 |
| serpinf1 | 0.206 | rnaseka | -0.030 |
| LOC100535907 | 0.205 | itga1 | -0.030 |
| vkorc1 | 0.200 | slc2a15a | -0.029 |
| col9a1a | 0.197 | mitfa | -0.029 |
| mmp14a | 0.194 | pah | -0.029 |
| ecm2 | 0.193 | qdpra | -0.029 |
| gpx7 | 0.192 | C12orf75 | -0.029 |
| LOC100536441 | 0.191 | aclya | -0.029 |
| matn4 | 0.186 | gstp1 | -0.029 |
| RNF224 | 0.185 | coro1a | -0.029 |
| prrx1a | 0.185 | rabl6b | -0.029 |
| ccl25b | 0.185 | trpm1b | -0.028 |
| lum | 0.182 | mibp | -0.028 |
| BX537274.2 | 0.181 | aldh16a1 | -0.027 |
| BX324006.1 | 0.181 | slc45a2 | -0.027 |
| mfap2 | 0.180 | rab3ip | -0.027 |
| srpx | 0.180 | pts | -0.027 |
| foxc1a | 0.178 | gch2 | -0.027 |
| mir214a | 0.176 | smim29 | -0.027 |