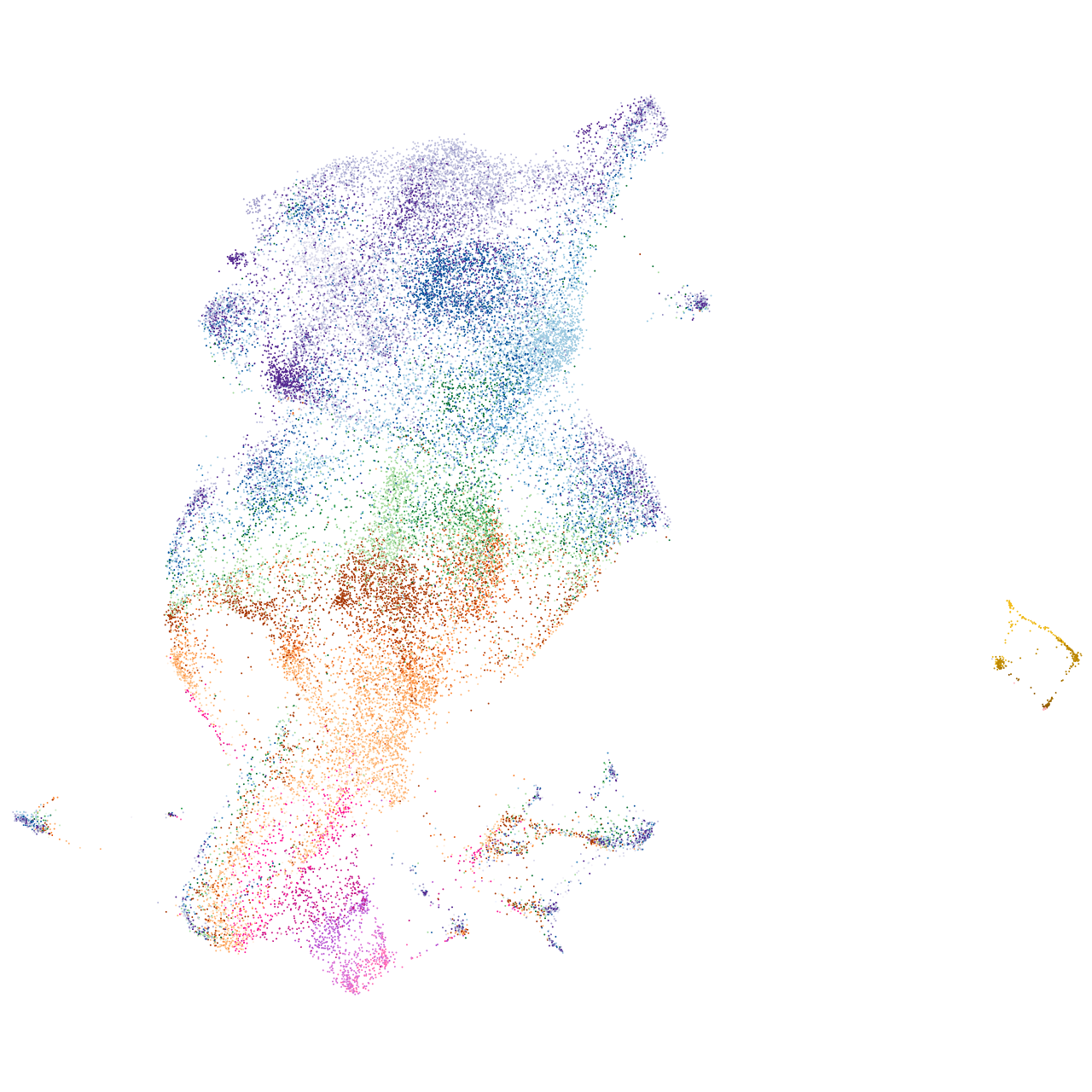SIX homeobox 1b
ZFIN 
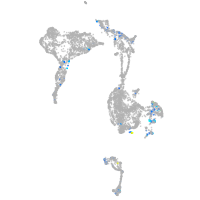
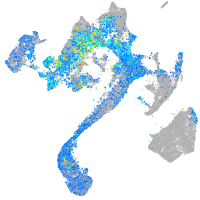
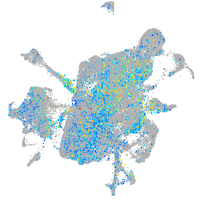
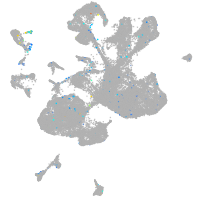
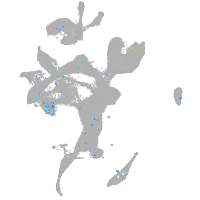
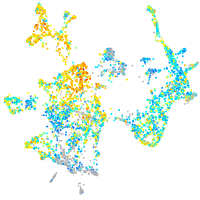
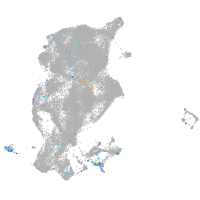
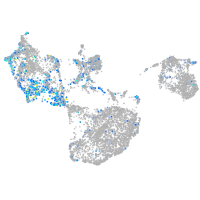
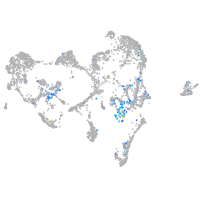
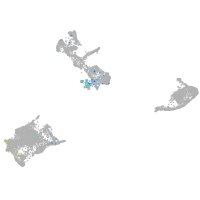
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| grp | 0.200 | si:ch211-195b11.3 | -0.066 |
| npy2r | 0.200 | cyt1 | -0.063 |
| CABZ01095001.1 | 0.192 | cfl1l | -0.058 |
| itga2.1 | 0.192 | wu:fb18f06 | -0.051 |
| CR759836.1 | 0.190 | krt4 | -0.049 |
| pvalb8 | 0.186 | anxa1a | -0.047 |
| dmrta2 | 0.185 | zgc:193505 | -0.044 |
| cldnh | 0.176 | agr1 | -0.042 |
| prr15la | 0.173 | si:ch73-52f15.5 | -0.041 |
| gfral | 0.172 | cyt1l | -0.041 |
| hcrtr2 | 0.170 | krt5 | -0.041 |
| CABZ01073083.1 | 0.168 | anxa1c | -0.040 |
| SCARF2 | 0.164 | scel | -0.039 |
| otofa | 0.159 | nqo1 | -0.037 |
| eya1 | 0.150 | aldob | -0.035 |
| si:ch211-256e16.6 | 0.148 | s100a10b | -0.035 |
| vill | 0.147 | si:dkey-16p21.8 | -0.034 |
| cers3b | 0.143 | si:ch211-157c3.4 | -0.034 |
| tph1b | 0.143 | pycard | -0.034 |
| stom | 0.141 | zgc:163030 | -0.034 |
| ret | 0.140 | ckap4 | -0.033 |
| six1a | 0.139 | eevs | -0.031 |
| hhla1 | 0.134 | selenow2b | -0.031 |
| cga | 0.131 | stx11b.1 | -0.031 |
| twist1a | 0.131 | smdt1b | -0.030 |
| stx2b | 0.131 | BX901957.1 | -0.030 |
| sox1b | 0.129 | eef1da | -0.029 |
| kcnn3 | 0.128 | fxyd6l | -0.029 |
| prrx1a | 0.124 | selenow1 | -0.029 |
| tbx4 | 0.122 | zgc:175088 | -0.029 |
| vwde | 0.121 | pfn1 | -0.028 |
| si:ch211-57i17.5 | 0.119 | tlcd1 | -0.028 |
| gper1 | 0.116 | apodb | -0.028 |
| hpcal1 | 0.116 | them6 | -0.028 |
| pth1a | 0.115 | krt17 | -0.028 |