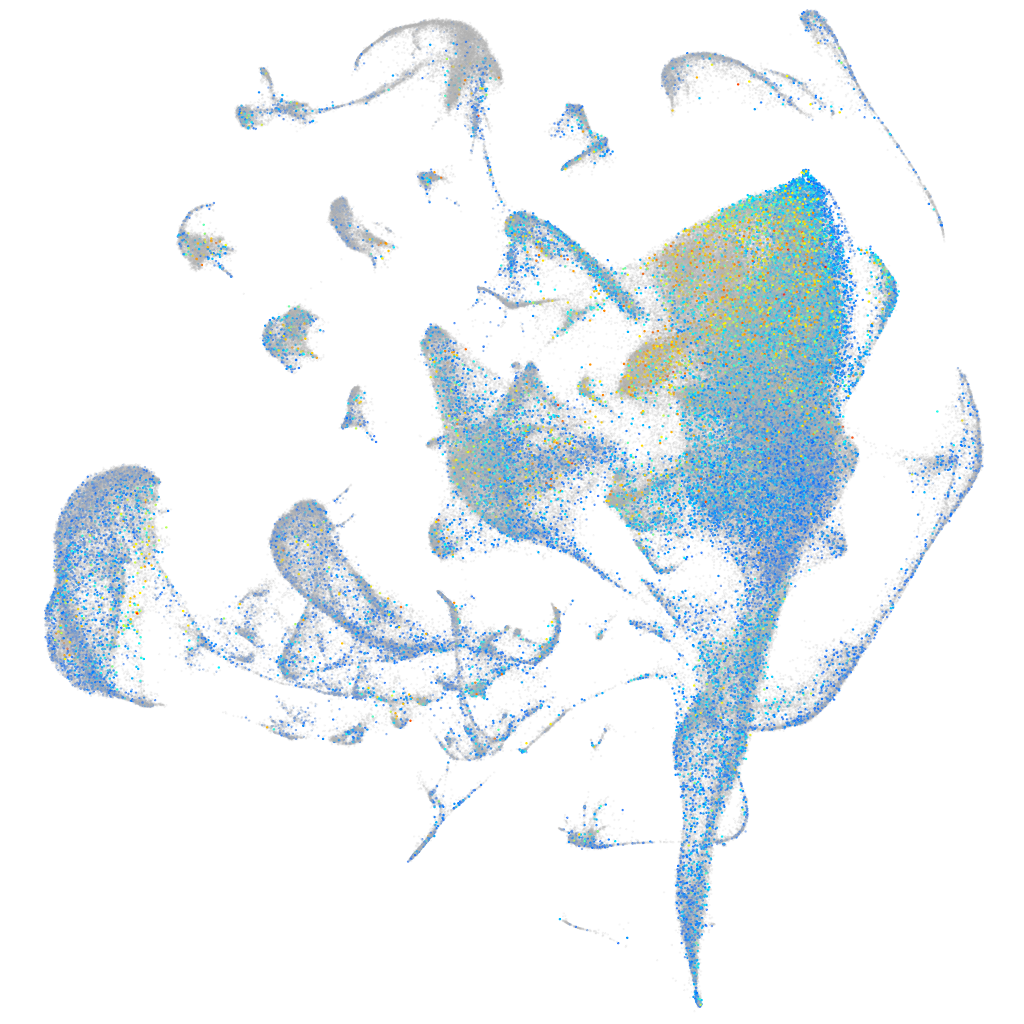si:dkeyp-118b1.2
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| elavl3 | 0.109 | si:dkey-16p21.8 | -0.047 |
| hmgb3a | 0.100 | aldob | -0.042 |
| tuba1c | 0.100 | eef1da | -0.042 |
| nova2 | 0.099 | alas2 | -0.040 |
| stmn1b | 0.097 | hbbe2 | -0.040 |
| tmeff1b | 0.096 | hemgn | -0.039 |
| si:dkey-276j7.1 | 0.094 | pfn1 | -0.039 |
| elavl4 | 0.093 | tuba8l2 | -0.039 |
| rtn1a | 0.093 | blvrb | -0.038 |
| snap25a | 0.092 | cahz | -0.038 |
| tubb5 | 0.091 | cebpd | -0.038 |
| stx1b | 0.090 | nt5c2l1 | -0.038 |
| ywhah | 0.090 | tspo | -0.038 |
| stxbp1a | 0.090 | zgc:162730 | -0.038 |
| gng3 | 0.088 | eno3 | -0.037 |
| zc4h2 | 0.088 | zgc:163057 | -0.037 |
| sncb | 0.087 | cfl1l | -0.036 |
| fez1 | 0.086 | epb41b | -0.036 |
| fam168a | 0.085 | gpx1a | -0.036 |
| myt1b | 0.085 | slc4a1a | -0.036 |
| tuba1a | 0.085 | tmod4 | -0.036 |
| vamp2 | 0.085 | ccng1 | -0.035 |
| dpysl2b | 0.085 | s100a10b | -0.035 |
| gnao1a | 0.084 | si:ch211-207c6.2 | -0.035 |
| tmsb | 0.084 | si:ch211-250g4.3 | -0.035 |
| dpysl3 | 0.083 | apoa2 | -0.034 |
| stmn2a | 0.083 | cebpb | -0.034 |
| gpm6ab | 0.082 | gamt | -0.034 |
| jpt1b | 0.082 | gapdh | -0.034 |
| gng2 | 0.081 | krt8 | -0.034 |
| mllt11 | 0.081 | nmt1b | -0.034 |
| gpm6aa | 0.080 | rbp4 | -0.034 |
| myt1a | 0.080 | anxa11a | -0.033 |
| ywhag2 | 0.080 | eif4ebp3l | -0.033 |
| zgc:65894 | 0.079 | gsta.1 | -0.033 |