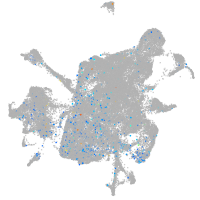
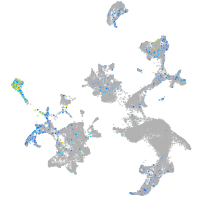
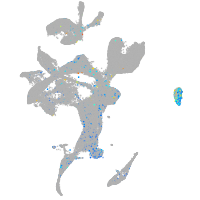
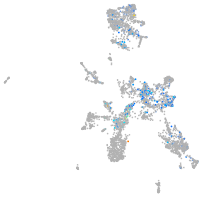
si:dkey-92i17.2
ZFIN 
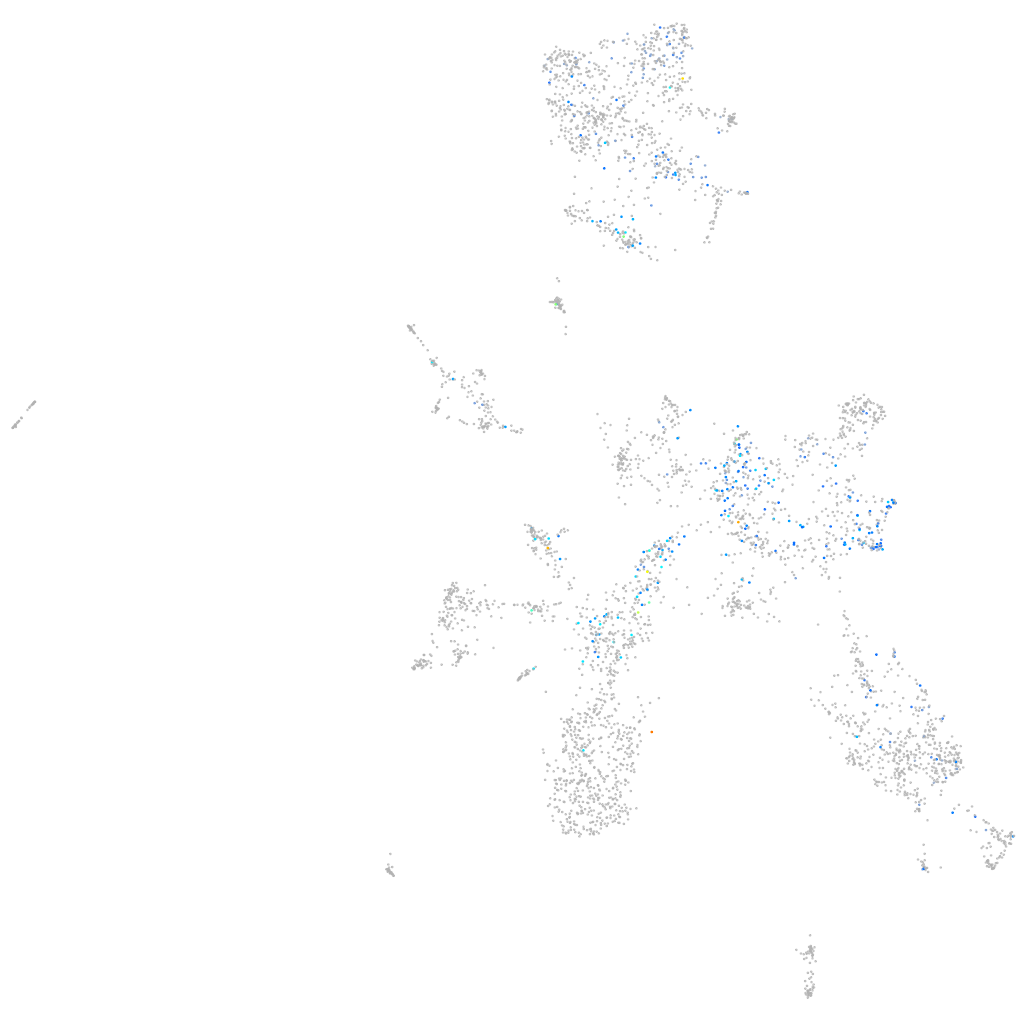
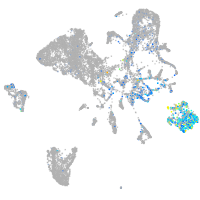
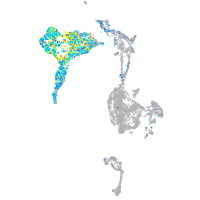
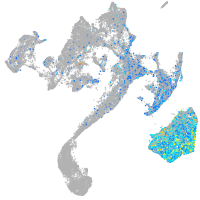
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dlx4b | 0.228 | gapdhs | -0.176 |
| hmga1a | 0.214 | gstp1 | -0.173 |
| hnrnpaba | 0.202 | selenow2b | -0.163 |
| h2afvb | 0.202 | txn | -0.147 |
| ddx39ab | 0.198 | fxyd1 | -0.139 |
| foxe3 | 0.192 | wu:fb18f06 | -0.138 |
| si:ch211-222l21.1 | 0.192 | si:ch211-153b23.5 | -0.135 |
| dlx3b | 0.191 | chga | -0.130 |
| ebf2 | 0.189 | nfe2l2a | -0.128 |
| XLOC-016803 | 0.188 | tmem59 | -0.127 |
| zmp:0000000912 | 0.187 | pvalb5 | -0.125 |
| fezf1 | 0.185 | tpi1b | -0.124 |
| CU467822.1 | 0.182 | pvalb1 | -0.122 |
| hmgb2b | 0.182 | fabp10b | -0.122 |
| her15.1 | 0.181 | icn | -0.122 |
| khdrbs1a | 0.180 | nqo1 | -0.118 |
| cxcr4b | 0.180 | si:dkey-71b5.7 | -0.118 |
| dlb | 0.175 | msi2b | -0.116 |
| ptmab | 0.174 | htatip2 | -0.115 |
| neurod4 | 0.174 | nrsn1 | -0.114 |
| neurod1 | 0.173 | eno1a | -0.113 |
| smarce1 | 0.173 | sod1 | -0.110 |
| rtca | 0.172 | gsto2 | -0.110 |
| znf1182 | 0.171 | map1aa | -0.108 |
| apex1 | 0.169 | gnb1a | -0.105 |
| nono | 0.169 | pvalb2 | -0.105 |
| emx3 | 0.169 | COX3 | -0.105 |
| cirbpa | 0.169 | anxa5b | -0.104 |
| h3f3a | 0.169 | pcsk1nl | -0.104 |
| snrpd2 | 0.168 | cplx2l | -0.103 |
| cdkn1ca | 0.168 | olfm1b | -0.103 |
| cirbpb | 0.167 | itm2ba | -0.102 |
| hmgn6 | 0.167 | flrt1b | -0.102 |
| hnrnpabb | 0.167 | TSTD1 | -0.102 |
| h2afva | 0.166 | kif1aa | -0.102 |