si:dkey-63j12.4
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 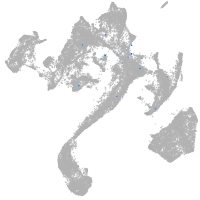 |
mural cells / non-skeletal muscle 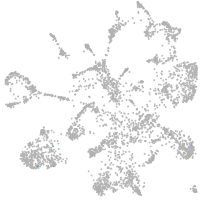 |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 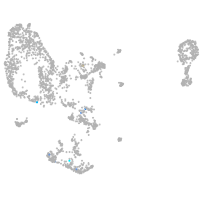 |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm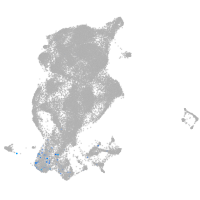 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:cabz01021433.2 | 0.184 | rtn1a | -0.031 |
| LOC110438082 | 0.157 | marcksl1b | -0.026 |
| spaw | 0.124 | jpt1b | -0.026 |
| ankdd1b | 0.116 | elavl3 | -0.025 |
| LOC110439434 | 0.108 | ckbb | -0.025 |
| si:dkey-26i13.5 | 0.103 | si:dkeyp-75h12.5 | -0.025 |
| XLOC-038091 | 0.099 | ywhah | -0.025 |
| LOC108181107 | 0.097 | rnasekb | -0.024 |
| BX323586.2 | 0.091 | gng3 | -0.023 |
| prp | 0.091 | calm1a | -0.023 |
| zgc:158366 | 0.075 | vamp2 | -0.023 |
| LOC110438598 | 0.075 | kdm6bb | -0.023 |
| BX890562.2 | 0.074 | sncb | -0.022 |
| si:ch1073-166e24.4 | 0.072 | stx1b | -0.022 |
| CT009620.1 | 0.068 | gng7 | -0.022 |
| si:ch211-146l10.7 | 0.067 | dpysl3 | -0.022 |
| zgc:110216 | 0.062 | syt2a | -0.021 |
| mibp | 0.060 | elavl4 | -0.021 |
| dut | 0.060 | tmsb | -0.021 |
| LOC101883798 | 0.060 | myt1b | -0.021 |
| ccna2 | 0.059 | mllt11 | -0.021 |
| mki67 | 0.059 | nsg2 | -0.021 |
| zgc:110540 | 0.059 | ppdpfb | -0.021 |
| pcna | 0.059 | stxbp1a | -0.021 |
| pidd1 | 0.059 | gnao1a | -0.020 |
| rrm2 | 0.059 | snap25a | -0.020 |
| ncaph | 0.059 | atp6v1e1b | -0.020 |
| fbxo5 | 0.058 | wu:fb55g09 | -0.020 |
| lig1 | 0.058 | rtn1b | -0.020 |
| ncapg | 0.058 | cplx2 | -0.020 |
| AL929536.3 | 0.058 | ywhag2 | -0.020 |
| esco2 | 0.058 | calm2a | -0.020 |
| rrm1 | 0.057 | stmn2a | -0.020 |
| cdca5 | 0.057 | scrt2 | -0.019 |
| mis18a | 0.056 | csdc2a | -0.019 |




