si:dkey-57k2.6
ZFIN 
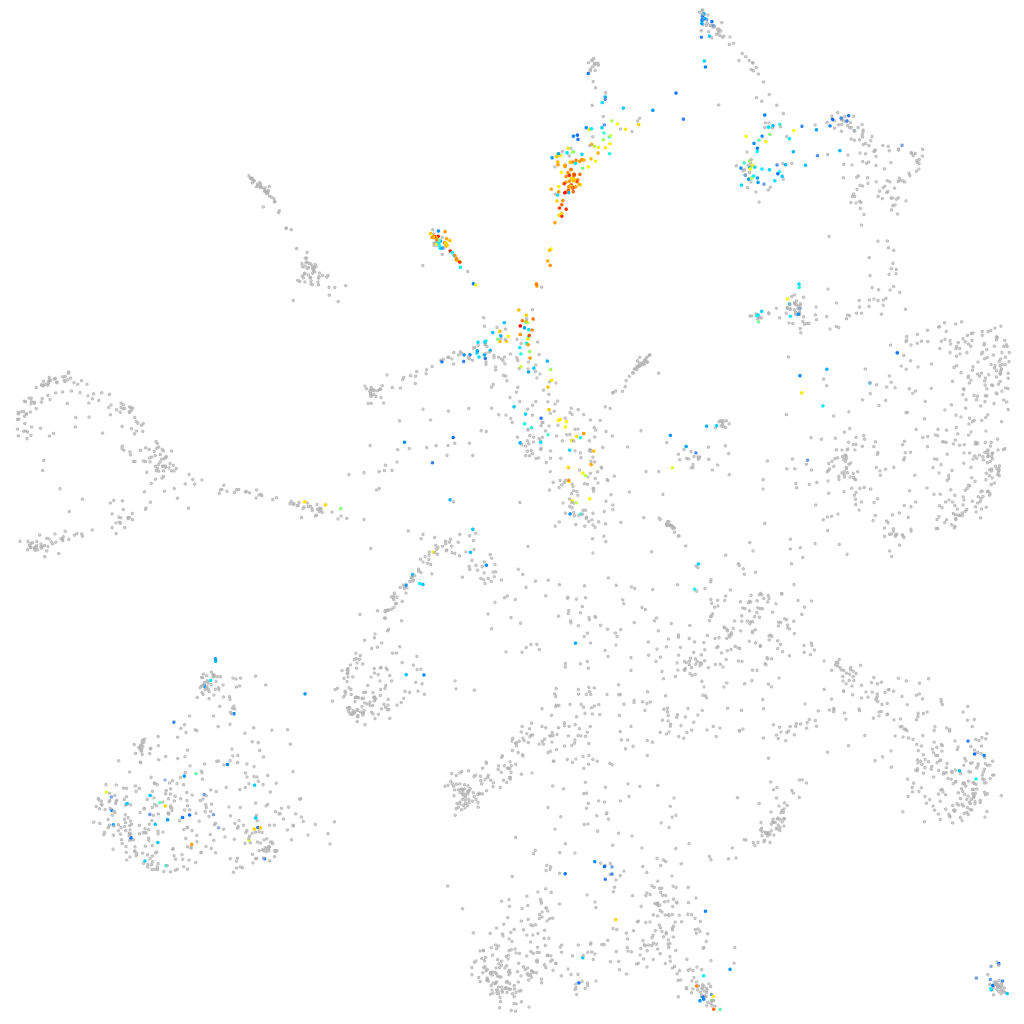
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| fbln5 | 0.559 | hmga2 | -0.192 |
| htra1a | 0.534 | foxf1 | -0.182 |
| loxa | 0.517 | nkx2.3 | -0.182 |
| si:ch211-1a19.3 | 0.494 | hlx1 | -0.171 |
| elnb | 0.489 | si:dkey-56m19.5 | -0.166 |
| BX663503.3 | 0.489 | col6a2 | -0.165 |
| si:dkey-164f24.2 | 0.471 | prdx1 | -0.159 |
| vim | 0.400 | si:dkeyp-66d1.7 | -0.159 |
| CABZ01092746.1 | 0.396 | col6a1 | -0.157 |
| c7a | 0.352 | ntn5 | -0.157 |
| acana | 0.342 | foxf2a | -0.157 |
| cyp2ad2 | 0.342 | XLOC-041870 | -0.152 |
| crip1 | 0.342 | si:ch211-62a1.3 | -0.147 |
| dub | 0.341 | col11a1a | -0.146 |
| emid1 | 0.330 | ckbb | -0.144 |
| crip3 | 0.328 | col4a5 | -0.144 |
| hpgd | 0.325 | col4a6 | -0.143 |
| s1pr3a | 0.319 | hmga1a | -0.140 |
| FP102018.1 | 0.311 | mylkb | -0.134 |
| fhl1b | 0.299 | marcksl1a | -0.134 |
| gpr78a | 0.299 | pdgfra | -0.132 |
| tnfsf12 | 0.290 | hmgb3a | -0.132 |
| wfdc2 | 0.289 | foxp4 | -0.130 |
| lmcd1 | 0.289 | ndnf | -0.128 |
| CR848683.2 | 0.286 | zgc:153704 | -0.125 |
| dapk3 | 0.284 | nkx3.3 | -0.124 |
| filip1b | 0.275 | fabp3 | -0.121 |
| crispld1b | 0.273 | tuba8l2 | -0.120 |
| tpm4b | 0.272 | inhbaa | -0.120 |
| angptl2a | 0.264 | meis1a | -0.116 |
| foxc1a | 0.263 | csrp1b | -0.112 |
| loxl1 | 0.253 | mab21l2 | -0.111 |
| ifitm1 | 0.253 | snrpd1 | -0.111 |
| mxra8b | 0.252 | hoxc8a | -0.110 |
| dap1b | 0.246 | bmp16 | -0.110 |