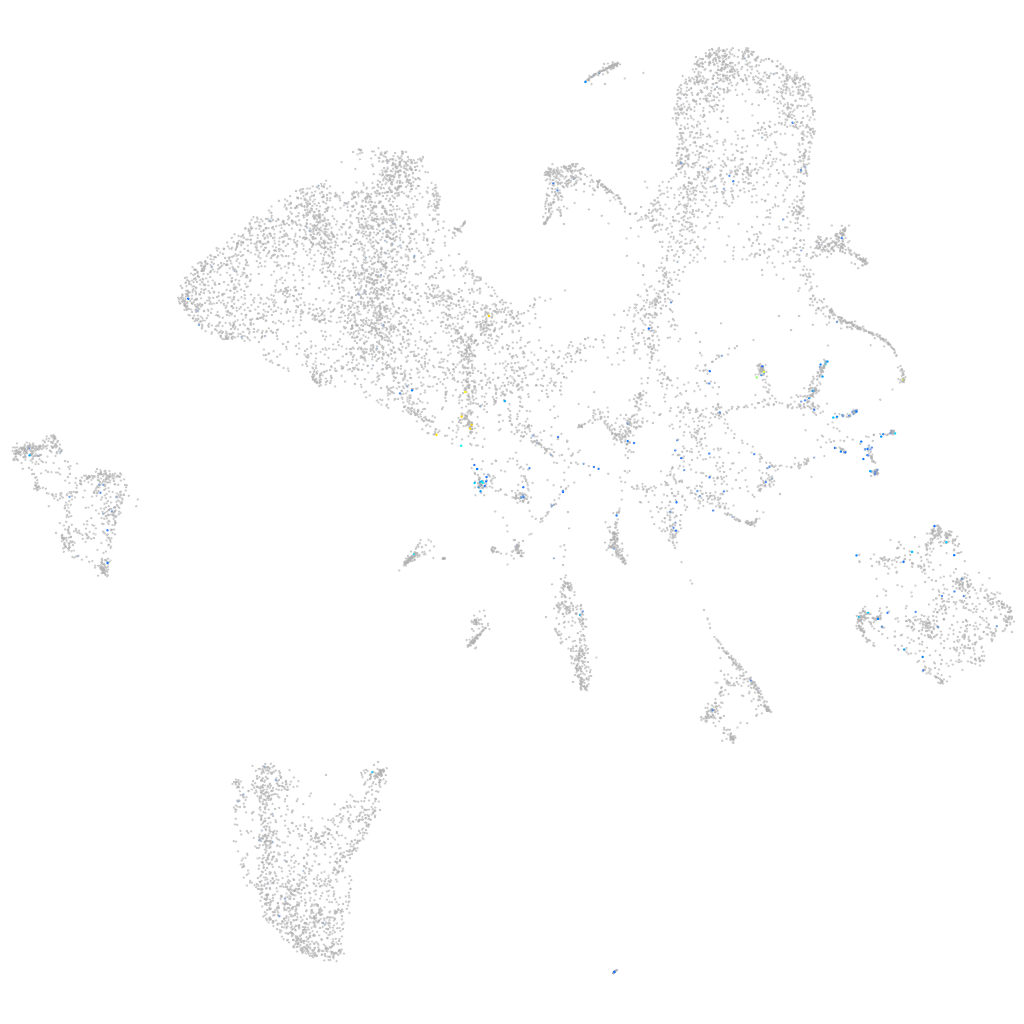
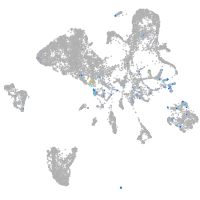
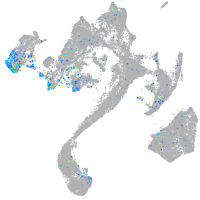
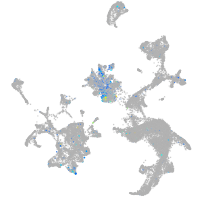
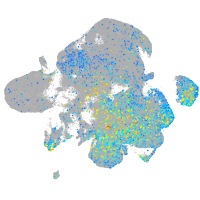
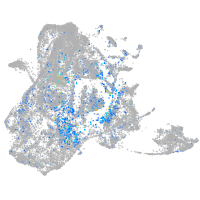
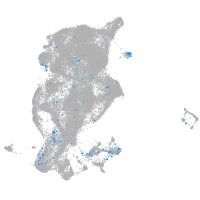
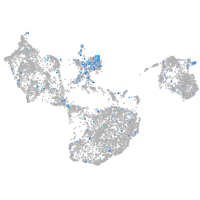
si:dkey-46i9.1
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cnrip1a | 0.226 | ahcy | -0.101 |
| myo1hb | 0.217 | gapdh | -0.098 |
| kcnc3a | 0.208 | aldob | -0.096 |
| si:ch73-287m6.1 | 0.206 | gamt | -0.087 |
| syt1a | 0.205 | dap | -0.081 |
| aldoca | 0.201 | nupr1b | -0.081 |
| tmem255a | 0.200 | eno3 | -0.080 |
| si:ch211-117c9.1 | 0.198 | aldh7a1 | -0.079 |
| CABZ01118767.1 | 0.193 | fbp1b | -0.078 |
| pcsk1 | 0.192 | rmdn1 | -0.077 |
| gng3 | 0.191 | cx32.3 | -0.077 |
| olfm1a | 0.189 | eef1da | -0.077 |
| gabrb4 | 0.186 | glud1b | -0.076 |
| slitrk3a | 0.185 | gatm | -0.075 |
| XLOC-022269 | 0.182 | hdlbpa | -0.075 |
| col28a2b | 0.182 | gstr | -0.075 |
| XLOC-029934 | 0.180 | apoa4b.1 | -0.074 |
| scgn | 0.180 | acadm | -0.073 |
| slc8a4a | 0.179 | aldh9a1a.1 | -0.072 |
| scg2b | 0.177 | mat1a | -0.072 |
| gpr17 | 0.177 | prdx6 | -0.072 |
| nmur3 | 0.177 | gcshb | -0.072 |
| diras1b | 0.177 | gstt1a | -0.072 |
| shank1 | 0.175 | scp2a | -0.071 |
| LO018020.2 | 0.174 | ckba | -0.070 |
| add2 | 0.174 | gstp1 | -0.069 |
| vamp2 | 0.172 | sod2 | -0.068 |
| stx1b | 0.172 | suclg1 | -0.068 |
| stxbp1a | 0.171 | gpx4a | -0.067 |
| elavl4 | 0.170 | si:dkey-16p21.8 | -0.067 |
| sv2a | 0.170 | aldh1l1 | -0.067 |
| kcnk15 | 0.169 | acmsd | -0.066 |
| si:dkey-70p6.1 | 0.169 | akt2l | -0.066 |
| snap25a | 0.168 | apoc2 | -0.066 |
| gnao1a | 0.168 | suclg2 | -0.066 |