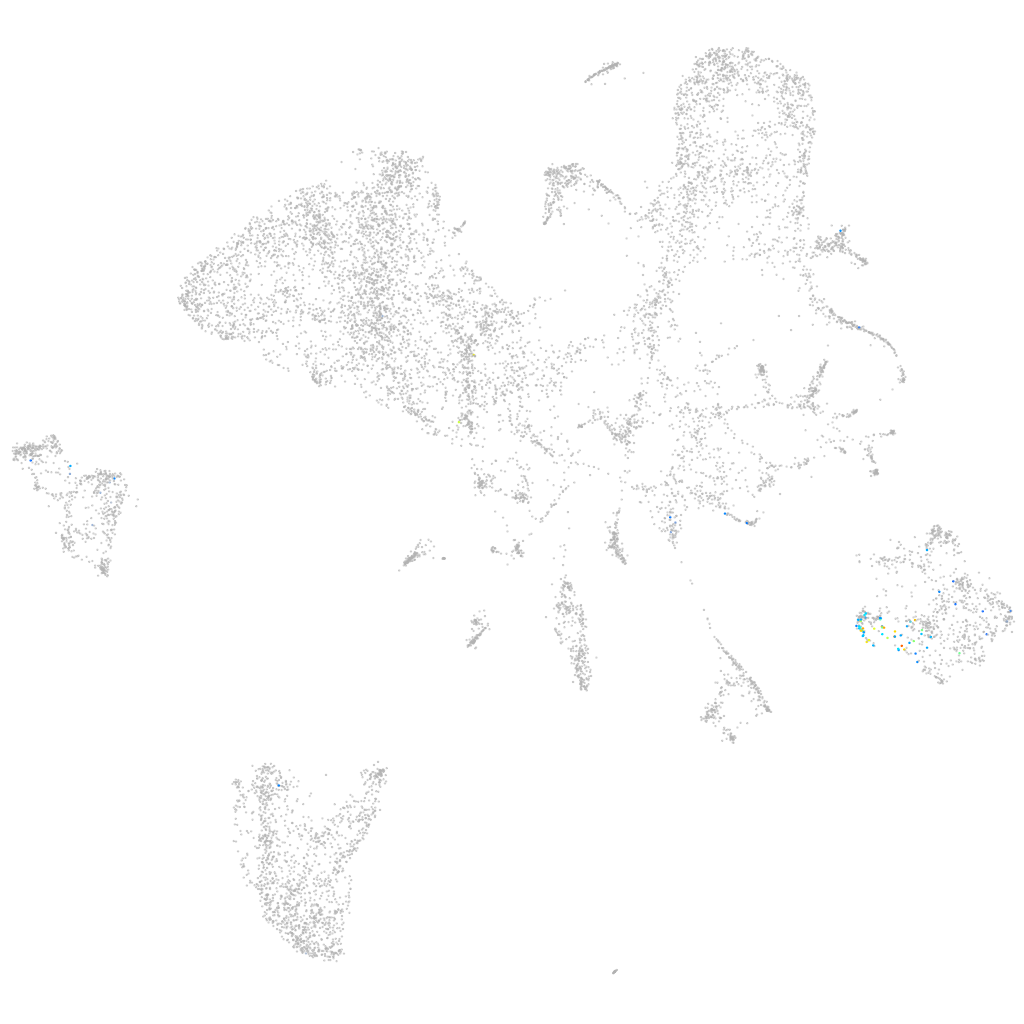
si:dkey-43p13.5
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle 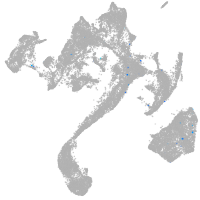 |
mural cells / non-skeletal muscle  |
mesenchyme 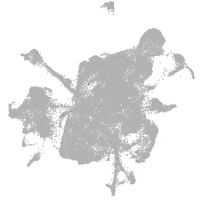 |
hematopoietic / vasculature 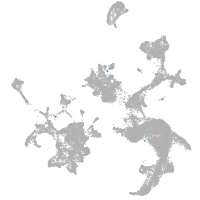 |
pronephros 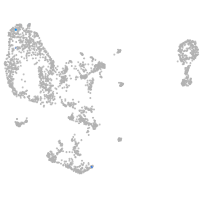 |
neural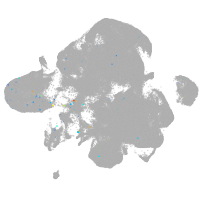 |
spinal cord / glia |
eye |
taste / olfactory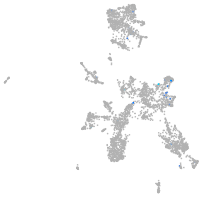 |
otic / lateral line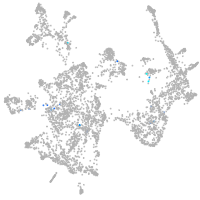 |
epidermis |
periderm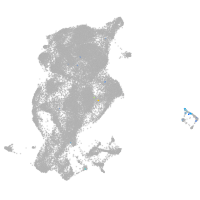 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cyp26c1 | 0.423 | rps10 | -0.185 |
| dmbx1a | 0.337 | rpl37 | -0.183 |
| otx1 | 0.304 | zgc:114188 | -0.171 |
| cxcl12b | 0.285 | rps17 | -0.158 |
| mcama | 0.267 | nme2b.1 | -0.118 |
| iqca1 | 0.253 | zgc:56493 | -0.106 |
| irx7 | 0.240 | atp5fa1 | -0.103 |
| cox4i2 | 0.236 | ahcy | -0.102 |
| otx2b | 0.230 | atp5l | -0.098 |
| akap12b | 0.218 | zgc:92744 | -0.097 |
| pltp | 0.217 | atp5mc3b | -0.097 |
| pax1a | 0.215 | prdx2 | -0.097 |
| asph | 0.214 | zgc:158463 | -0.096 |
| si:ch211-152c2.3 | 0.213 | atp5f1b | -0.095 |
| lima1a | 0.213 | atp5pf | -0.092 |
| gsc | 0.212 | mt-nd1 | -0.091 |
| zgc:162707 | 0.203 | atp5mc1 | -0.091 |
| BX649556.1 | 0.198 | sod1 | -0.090 |
| smoc2 | 0.197 | eno3 | -0.089 |
| CU571091.1 | 0.197 | eef1da | -0.089 |
| sh2d4ba | 0.191 | atp5if1b | -0.089 |
| arhgap36 | 0.190 | gapdh | -0.087 |
| LOC101886005 | 0.188 | atp5meb | -0.087 |
| nkx2.7 | 0.186 | COX7A2 (1 of many) | -0.086 |
| dbx1a | 0.185 | cox6a1 | -0.085 |
| cdh6 | 0.182 | suclg1 | -0.085 |
| efna1a | 0.178 | romo1 | -0.085 |
| BX649529.1 | 0.175 | cct2 | -0.083 |
| coro1cb | 0.172 | tpi1b | -0.081 |
| NC-002333.4 | 0.171 | gamt | -0.081 |
| crtap | 0.169 | gstp1 | -0.080 |
| gpm6ab | 0.169 | mt-atp6 | -0.079 |
| fscn2b | 0.168 | cox6b1 | -0.079 |
| rarga | 0.167 | hnrnpa0l | -0.079 |
| marcksl1b | 0.167 | ndufa3 | -0.079 |