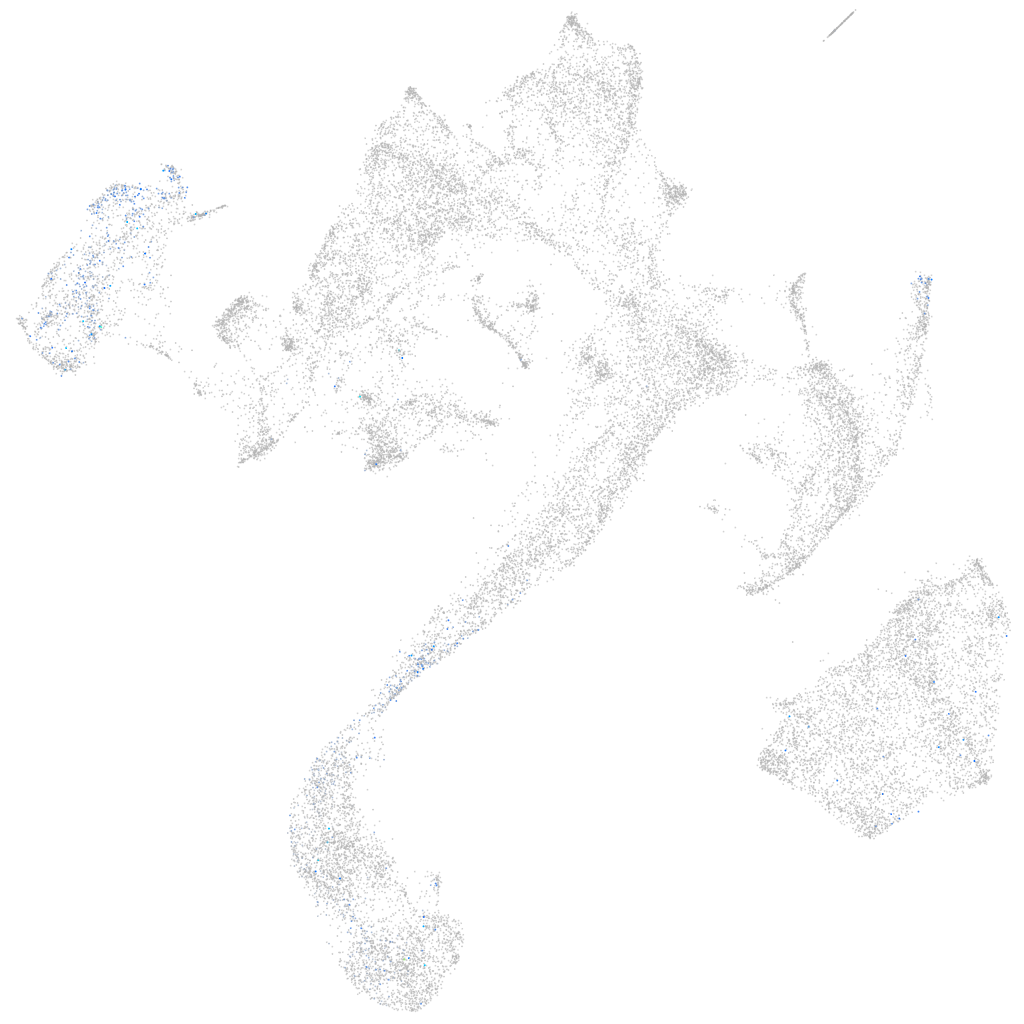
si:dkey-32e6.6
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm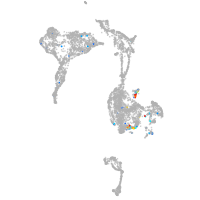 |
muscle 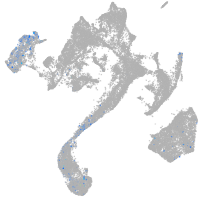 |
mural cells / non-skeletal muscle 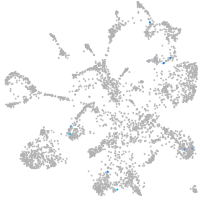 |
mesenchyme 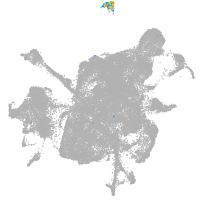 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:158296 | 0.231 | hsp90ab1 | -0.164 |
| tnnt2c | 0.224 | pabpc1a | -0.153 |
| smyhc1 | 0.222 | hmgb2b | -0.152 |
| tnnc1b | 0.221 | ran | -0.147 |
| myl13 | 0.219 | si:ch73-1a9.3 | -0.146 |
| srl | 0.218 | ubc | -0.139 |
| CU633479.2 | 0.218 | si:ch211-222l21.1 | -0.137 |
| tnni1c | 0.214 | khdrbs1a | -0.137 |
| casq2 | 0.211 | hmgn2 | -0.135 |
| aldoab | 0.209 | h2afvb | -0.133 |
| LOC103910269 | 0.209 | hnrnpabb | -0.133 |
| cavin4b | 0.205 | hnrnpaba | -0.132 |
| myom1b | 0.204 | cirbpb | -0.132 |
| desma | 0.202 | ptmab | -0.132 |
| cyp4t8 | 0.201 | cirbpa | -0.132 |
| gapdh | 0.201 | h3f3d | -0.131 |
| myl10 | 0.201 | cfl1 | -0.130 |
| acta1a | 0.201 | hmgb2a | -0.126 |
| cav3 | 0.199 | tuba8l4 | -0.125 |
| parvb | 0.198 | hmgn7 | -0.125 |
| tpm2 | 0.198 | actb1 | -0.124 |
| trdn | 0.197 | hnrnpa0b | -0.124 |
| pgam2 | 0.197 | setb | -0.123 |
| eno3 | 0.196 | cbx3a | -0.121 |
| ryr1a | 0.196 | actb2 | -0.121 |
| zgc:86709 | 0.195 | si:ch73-281n10.2 | -0.120 |
| sar1ab | 0.195 | sumo3a | -0.118 |
| apobec2a | 0.195 | anp32b | -0.118 |
| LO018250.1 | 0.194 | snrpd1 | -0.118 |
| CABZ01072309.1 | 0.193 | snrpf | -0.118 |
| smtnl1 | 0.192 | h2afva | -0.117 |
| ldb3b | 0.191 | ranbp1 | -0.115 |
| tmem182a | 0.191 | tmsb4x | -0.115 |
| tnni4b.2 | 0.191 | tubb4b | -0.114 |
| actn3a | 0.190 | tubb2b | -0.114 |