si:dkey-261m9.17
ZFIN 
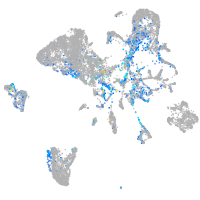
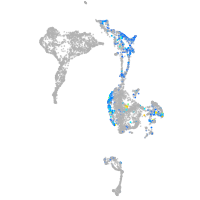
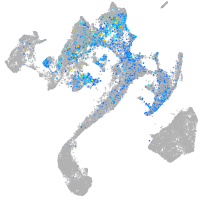
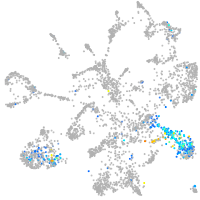
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:165555.3 | 0.619 | aldoab | -0.191 |
| hist1h4l | 0.522 | gapdh | -0.191 |
| zgc:153409 | 0.473 | ak1 | -0.185 |
| rrm2 | 0.434 | tmem38a | -0.182 |
| dut | 0.432 | tnnc2 | -0.182 |
| h3f3a | 0.419 | atp2a1 | -0.180 |
| tuba8l | 0.409 | ckma | -0.179 |
| hist1h2a11 | 0.394 | neb | -0.179 |
| cks1b | 0.375 | ldb3a | -0.177 |
| cks2 | 0.361 | ckmb | -0.177 |
| tubb2b | 0.356 | actc1b | -0.177 |
| tubb4b | 0.350 | srl | -0.176 |
| LOC100330864 | 0.341 | mybphb | -0.174 |
| stmn1a | 0.327 | ttn.1 | -0.174 |
| lbr | 0.325 | acta1b | -0.173 |
| tuba8l4 | 0.322 | ldb3b | -0.169 |
| cdk1 | 0.319 | actn3b | -0.169 |
| rrm1 | 0.313 | ttn.2 | -0.168 |
| si:dkey-6i22.5 | 0.310 | cav3 | -0.167 |
| iqgap3 | 0.306 | actn3a | -0.167 |
| hmgn2 | 0.305 | si:ch73-367p23.2 | -0.167 |
| ptmaa | 0.292 | desma | -0.166 |
| ccna2 | 0.291 | CABZ01078594.1 | -0.166 |
| aurkb | 0.291 | acta1a | -0.165 |
| CABZ01058261.1 | 0.290 | tpma | -0.164 |
| top2a | 0.290 | casq2 | -0.162 |
| sinhcafl | 0.288 | hhatla | -0.161 |
| ncapg | 0.286 | myom1a | -0.161 |
| cdca5 | 0.285 | smyd1a | -0.160 |
| zgc:56493 | 0.283 | si:ch211-266g18.10 | -0.160 |
| prim2 | 0.282 | nme2b.2 | -0.158 |
| pcna | 0.279 | tmod4 | -0.157 |
| trip13 | 0.278 | ank1a | -0.157 |
| hist1h2a6 | 0.276 | pgam2 | -0.156 |
| h2afx | 0.272 | trdn | -0.156 |