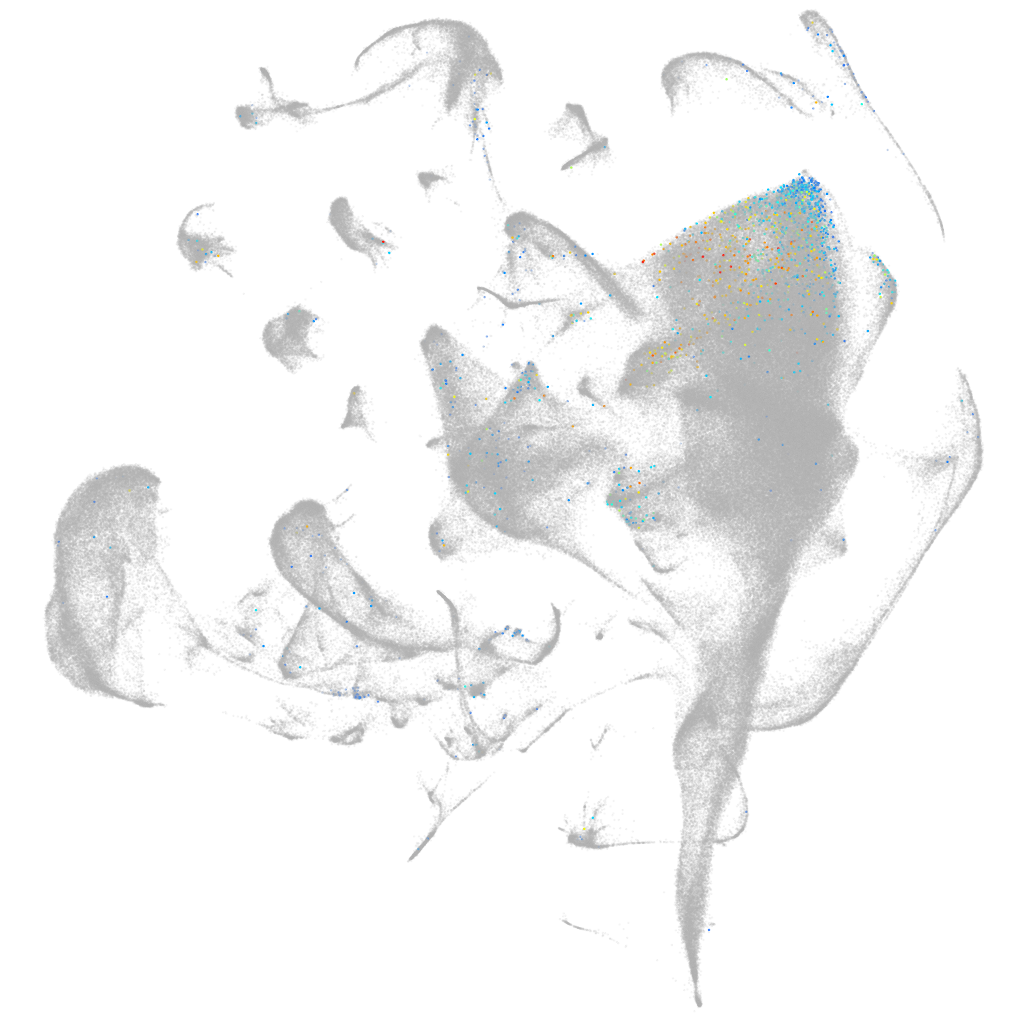si:dkey-192j17.1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 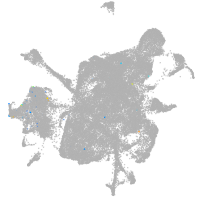 |
hematopoietic / vasculature 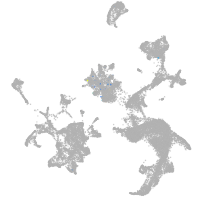 |
pronephros  |
neural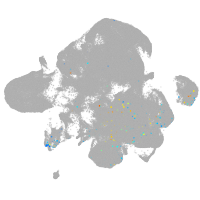 |
spinal cord / glia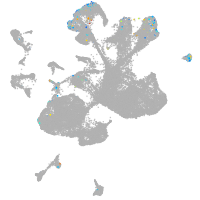 |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sncgb | 0.093 | hspb1 | -0.031 |
| eno1a | 0.088 | cx43.4 | -0.030 |
| aldocb | 0.085 | pou5f3 | -0.030 |
| tubb2 | 0.084 | aldob | -0.028 |
| snap25a | 0.083 | stm | -0.028 |
| cplx2 | 0.082 | arf1 | -0.027 |
| vamp1 | 0.082 | polr3gla | -0.027 |
| cplx2l | 0.081 | apoeb | -0.026 |
| calm1b | 0.079 | asb11 | -0.026 |
| sh3gl2a | 0.079 | nnr | -0.026 |
| stmn3 | 0.079 | si:ch1073-80i24.3 | -0.026 |
| atp2b3a | 0.078 | si:dkey-66i24.9 | -0.026 |
| atp6v0cb | 0.078 | apoc1 | -0.025 |
| scg2a | 0.078 | dkc1 | -0.025 |
| stxbp1a | 0.078 | fbl | -0.025 |
| ndufa4 | 0.077 | hdlbpa | -0.025 |
| pkd2l1 | 0.077 | nop58 | -0.025 |
| gapdhs | 0.076 | tbx16 | -0.025 |
| map6d1 | 0.076 | zgc:56699 | -0.025 |
| eno2 | 0.075 | bzw1b | -0.024 |
| stx1b | 0.075 | hnrnpa1b | -0.024 |
| atp1b2a | 0.074 | lig1 | -0.024 |
| map1aa | 0.074 | mki67 | -0.024 |
| pcsk1nl | 0.074 | npm1a | -0.024 |
| si:ch211-232m10.6 | 0.074 | tpx2 | -0.024 |
| sypb | 0.074 | ved | -0.024 |
| nptna | 0.073 | vox | -0.024 |
| ywhag2 | 0.073 | banf1 | -0.023 |
| atp2b3b | 0.072 | bms1 | -0.023 |
| syt2a | 0.072 | chaf1a | -0.023 |
| atp1a3b | 0.071 | dnmt3bb.2 | -0.023 |
| neflb | 0.071 | eif2s1a | -0.023 |
| nefmb | 0.071 | epcam | -0.023 |
| atpv0e2 | 0.070 | gar1 | -0.023 |
| si:ch73-119p20.1 | 0.070 | ing5b | -0.023 |