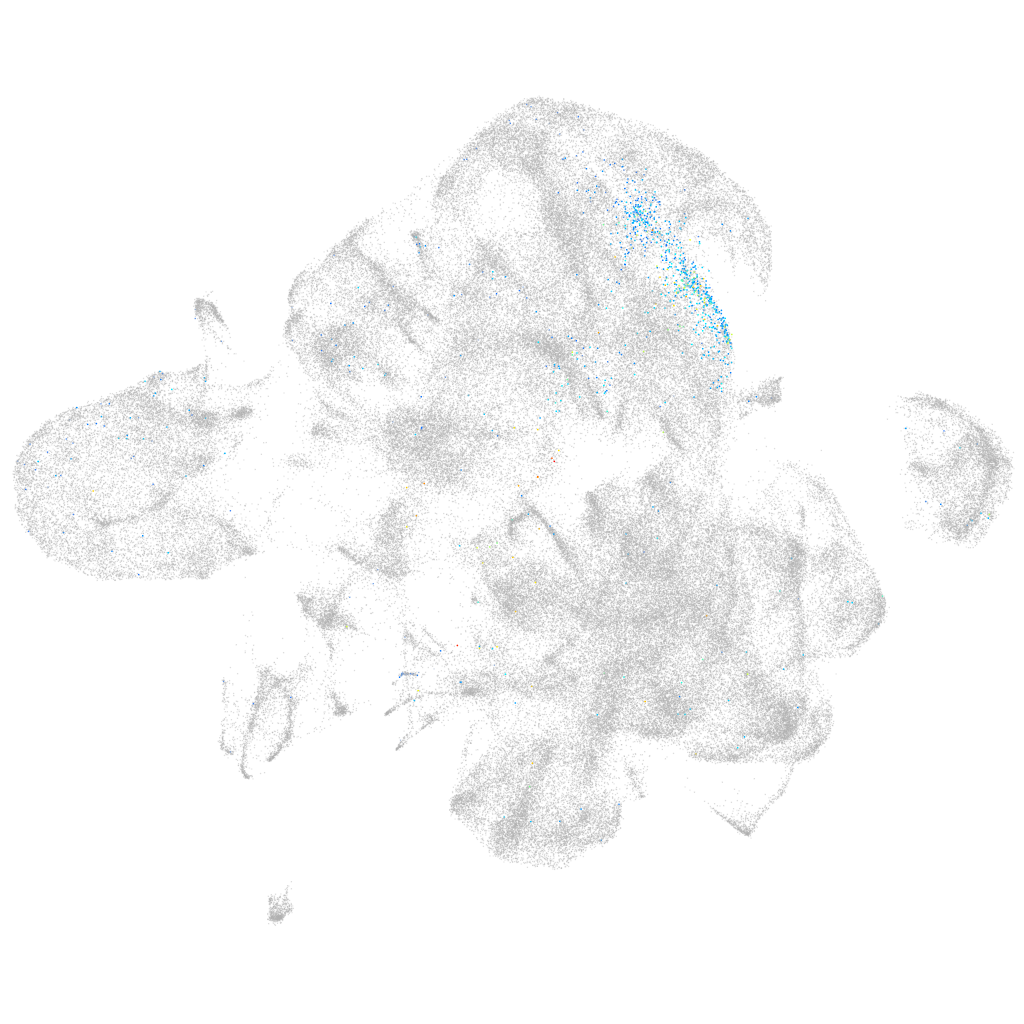
si:dkey-188g12.1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm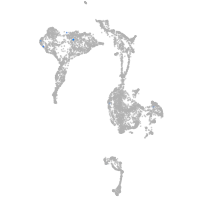 |
muscle 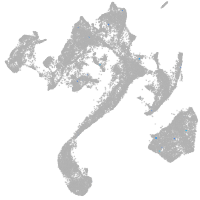 |
mural cells / non-skeletal muscle  |
mesenchyme 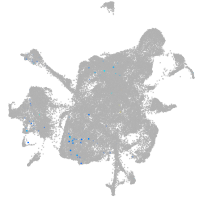 |
hematopoietic / vasculature 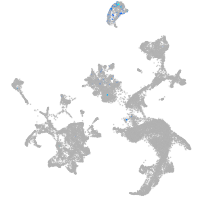 |
pronephros 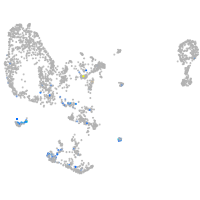 |
neural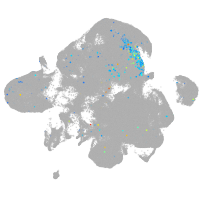 |
spinal cord / glia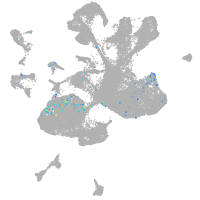 |
eye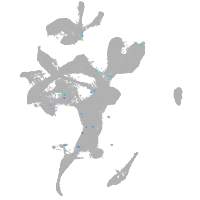 |
taste / olfactory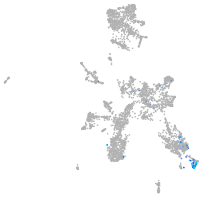 |
otic / lateral line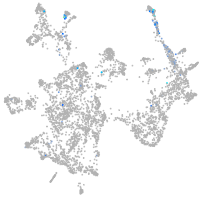 |
epidermis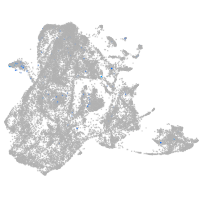 |
periderm |
fin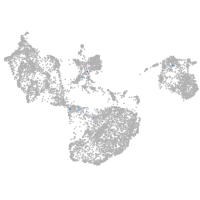 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| AL845362.1 | 0.440 | gng3 | -0.039 |
| ptf1a | 0.315 | h2afy2 | -0.038 |
| prdm13 | 0.196 | rtn1b | -0.036 |
| ascl1a | 0.183 | sncb | -0.035 |
| kirrel3l | 0.172 | elavl4 | -0.034 |
| lbx1b | 0.166 | calm2a | -0.034 |
| prss35 | 0.116 | stmn2a | -0.032 |
| gsx2 | 0.110 | atp6v0cb | -0.032 |
| lbx1a | 0.102 | atp6v1e1b | -0.032 |
| slc1a3a | 0.101 | stmn1b | -0.031 |
| gadd45gb.1 | 0.101 | ywhaz | -0.030 |
| lfng | 0.100 | stx1b | -0.030 |
| dld | 0.100 | snap25a | -0.029 |
| lrrn1 | 0.097 | ywhag2 | -0.029 |
| LOC798783 | 0.097 | gapdhs | -0.029 |
| dla | 0.096 | gpm6ab | -0.029 |
| fstl1a | 0.094 | tmem59l | -0.029 |
| si:ch211-251b21.1 | 0.092 | vamp2 | -0.029 |
| pax3a | 0.089 | tmsb2 | -0.029 |
| neurod4 | 0.086 | mllt11 | -0.029 |
| mdka | 0.085 | csdc2a | -0.029 |
| LOC101882472 | 0.085 | id4 | -0.028 |
| notch3 | 0.084 | gap43 | -0.028 |
| gpm6bb | 0.084 | stxbp1a | -0.028 |
| tfap2b | 0.084 | fez1 | -0.028 |
| wif1 | 0.083 | mab21l2 | -0.028 |
| zic1 | 0.082 | cdk5r1b | -0.028 |
| sertad2a | 0.081 | zgc:65894 | -0.028 |
| si:ch73-21g5.7 | 0.081 | gng2 | -0.028 |
| CR848812.1 | 0.080 | cbx1b | -0.028 |
| CU634008.1 | 0.078 | map1aa | -0.027 |
| plp1a | 0.078 | calm1a | -0.027 |
| smad3b | 0.077 | calm3b | -0.027 |
| cdkn1ca | 0.077 | olfm1b | -0.027 |
| her13 | 0.075 | cnrip1a | -0.026 |