si:ch73-81k8.2
ZFIN 
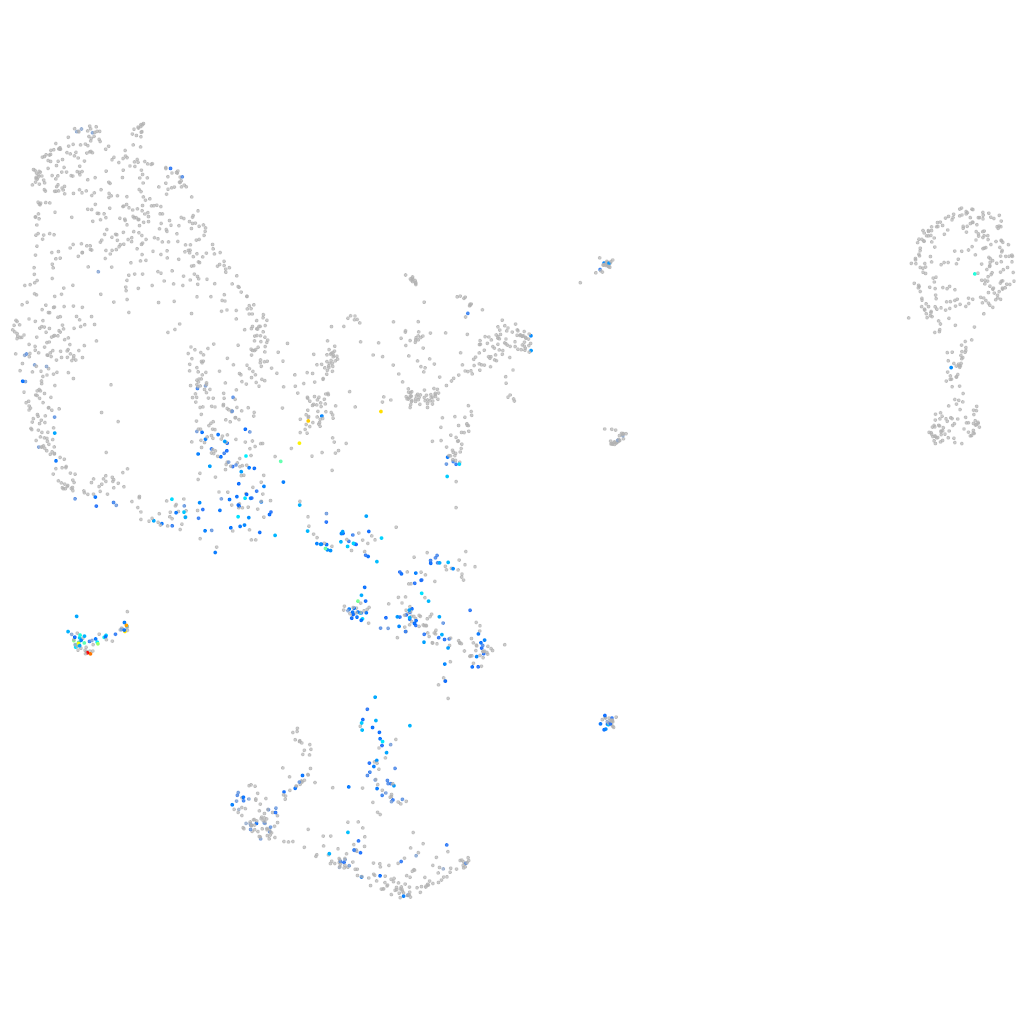
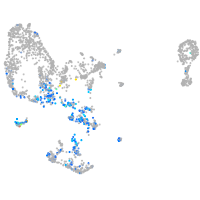
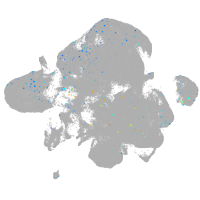
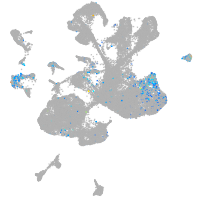
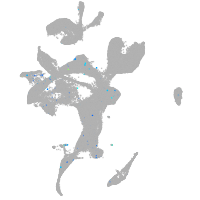
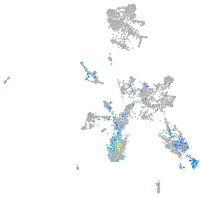
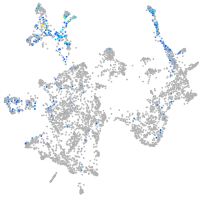
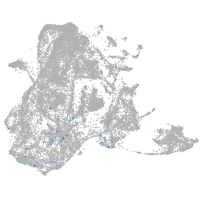
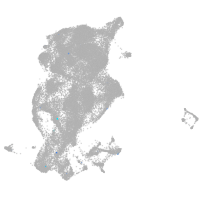
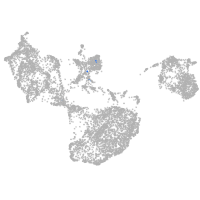
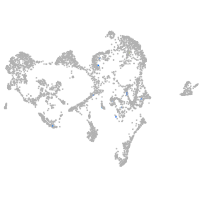
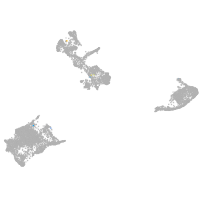
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:ch211-163l21.7 | 0.469 | gapdh | -0.216 |
| si:dkey-42p14.3 | 0.464 | gcshb | -0.208 |
| ttc25 | 0.462 | si:dkey-33i11.9 | -0.199 |
| capsla | 0.458 | alpl | -0.198 |
| iqcg | 0.457 | agxtb | -0.194 |
| ccdc114 | 0.447 | epdl2 | -0.192 |
| enkur | 0.445 | gpt2l | -0.186 |
| pacrg | 0.445 | si:ch211-139a5.9 | -0.186 |
| zgc:55461 | 0.443 | gstt1a | -0.183 |
| si:dkey-148f10.4 | 0.439 | hao1 | -0.181 |
| capslb | 0.432 | glud1b | -0.180 |
| tekt4 | 0.427 | cx32.3 | -0.179 |
| si:ch211-155m12.5 | 0.424 | phyhd1 | -0.179 |
| daw1 | 0.421 | dpys | -0.178 |
| mycbp | 0.418 | slc16a1b | -0.175 |
| si:dkeyp-46h3.1 | 0.413 | ASS1 | -0.174 |
| morn2 | 0.411 | cubn | -0.173 |
| cfap45 | 0.411 | nipsnap3a | -0.171 |
| efcab1 | 0.410 | g6pca.2 | -0.169 |
| tctex1d1 | 0.410 | lgmn | -0.168 |
| ccdc173 | 0.407 | aco1 | -0.168 |
| LOC103911624 | 0.407 | cox6a2 | -0.168 |
| cfap52 | 0.406 | rgn | -0.167 |
| tekt1 | 0.405 | pnp6 | -0.167 |
| si:ch211-248e11.2 | 0.405 | rps18 | -0.167 |
| spata4 | 0.403 | slc5a12 | -0.167 |
| meig1 | 0.400 | slc26a6l | -0.167 |
| ccdc169 | 0.399 | ctsz | -0.166 |
| smkr1 | 0.397 | fbp1b | -0.165 |
| rsph9 | 0.396 | pck2 | -0.164 |
| ribc2 | 0.392 | slc22a4 | -0.164 |
| zgc:195356 | 0.388 | rpl3 | -0.164 |
| spag8 | 0.387 | nme2b.1 | -0.163 |
| tbata | 0.387 | cat | -0.163 |
| spag6 | 0.387 | ctsla | -0.161 |