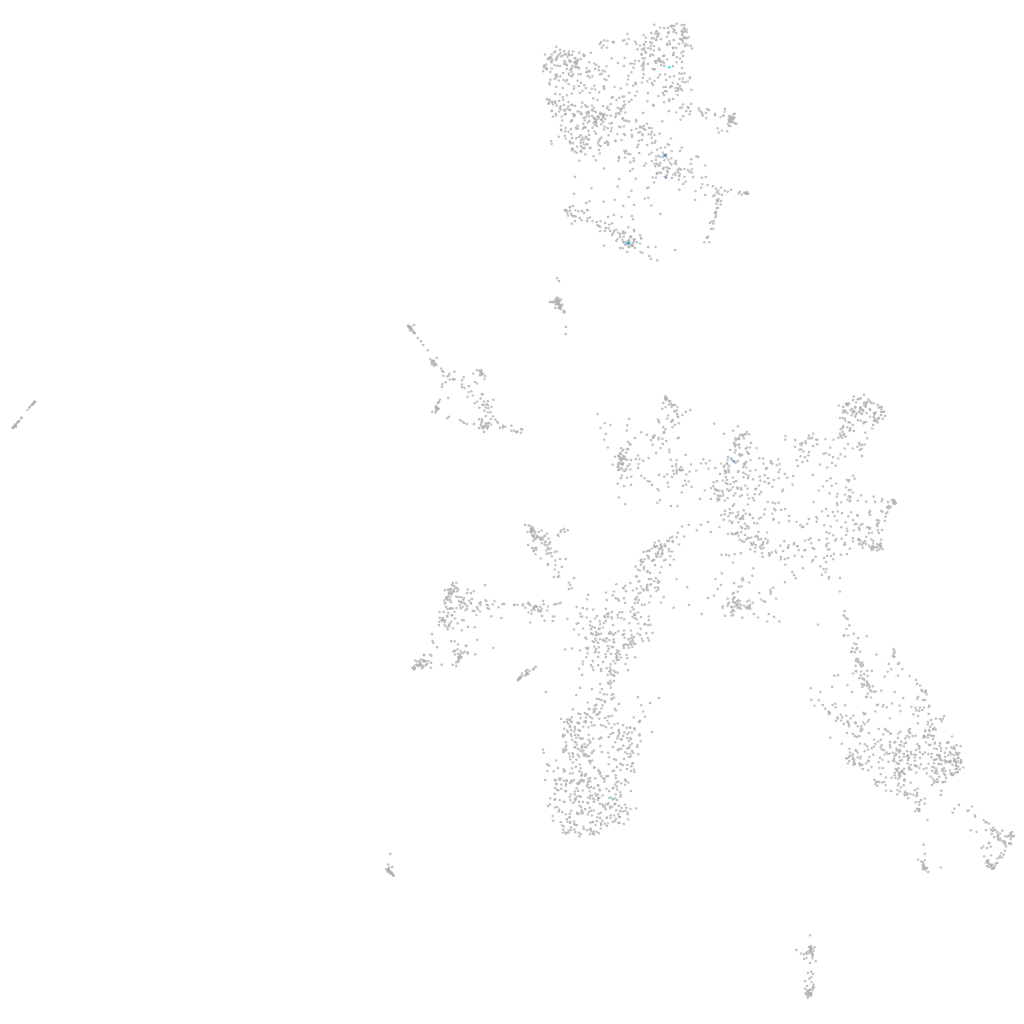
si:ch73-248e21.7
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 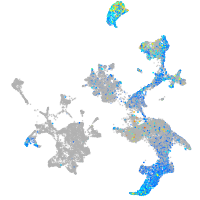 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory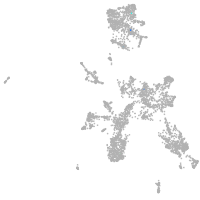 |
otic / lateral line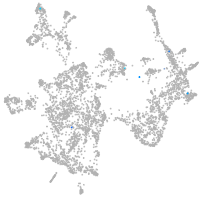 |
epidermis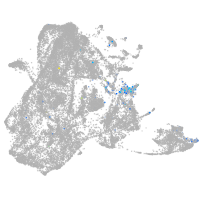 |
periderm |
fin |
ionocytes / mucous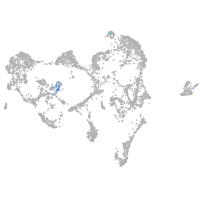 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01072815.1 | 0.584 | ndufb8 | -0.040 |
| adgrg3 | 0.567 | rpl19 | -0.039 |
| slc6a19b | 0.567 | gpx4b | -0.037 |
| zgc:172053 | 0.547 | ndufb3 | -0.037 |
| cdkn2a/b | 0.507 | sub1a | -0.037 |
| BX649522.1 | 0.462 | ywhae1 | -0.037 |
| znf1073 | 0.455 | vdac1 | -0.036 |
| myot | 0.410 | si:ch73-1a9.3 | -0.036 |
| ctss1 | 0.396 | ldhba | -0.036 |
| si:ch211-282j17.1 | 0.395 | atp5f1d | -0.036 |
| chrng | 0.393 | sumo2b | -0.035 |
| gda | 0.376 | rps12 | -0.035 |
| mustn1b | 0.354 | rps13 | -0.035 |
| parp6a | 0.339 | srsf5a | -0.035 |
| LOC108192030 | 0.327 | syncrip | -0.034 |
| bicd1a | 0.297 | f11r.1 | -0.034 |
| LOC108183685 | 0.294 | NC-002333.4 | -0.033 |
| CR394535.1 | 0.291 | cmpk | -0.033 |
| CR381686.5 | 0.286 | fth1a | -0.033 |
| sb:cb1058 | 0.284 | rps21 | -0.032 |
| CR450686.2 | 0.281 | dad1 | -0.032 |
| fer1l6 | 0.276 | atp5mc3a | -0.032 |
| CABZ01111953.1 | 0.273 | ssr2 | -0.032 |
| hs3st2 | 0.264 | rpl9 | -0.032 |
| slc51a | 0.252 | tegt | -0.032 |
| cpa5 | 0.244 | rpl35 | -0.031 |
| tnxba | 0.233 | rps15 | -0.031 |
| AL929028.1 | 0.230 | ppiab | -0.031 |
| XLOC-019883 | 0.225 | snrpd1 | -0.031 |
| LOC103911313 | 0.223 | COX5B | -0.031 |
| plekhn1 | 0.217 | morf4l1 | -0.031 |
| TMEM8B | 0.217 | cox6a1 | -0.030 |
| bach2a | 0.213 | marcksl1b | -0.030 |
| gnsb | 0.212 | ankrd11 | -0.030 |
| magixb | 0.211 | hmgn3 | -0.030 |