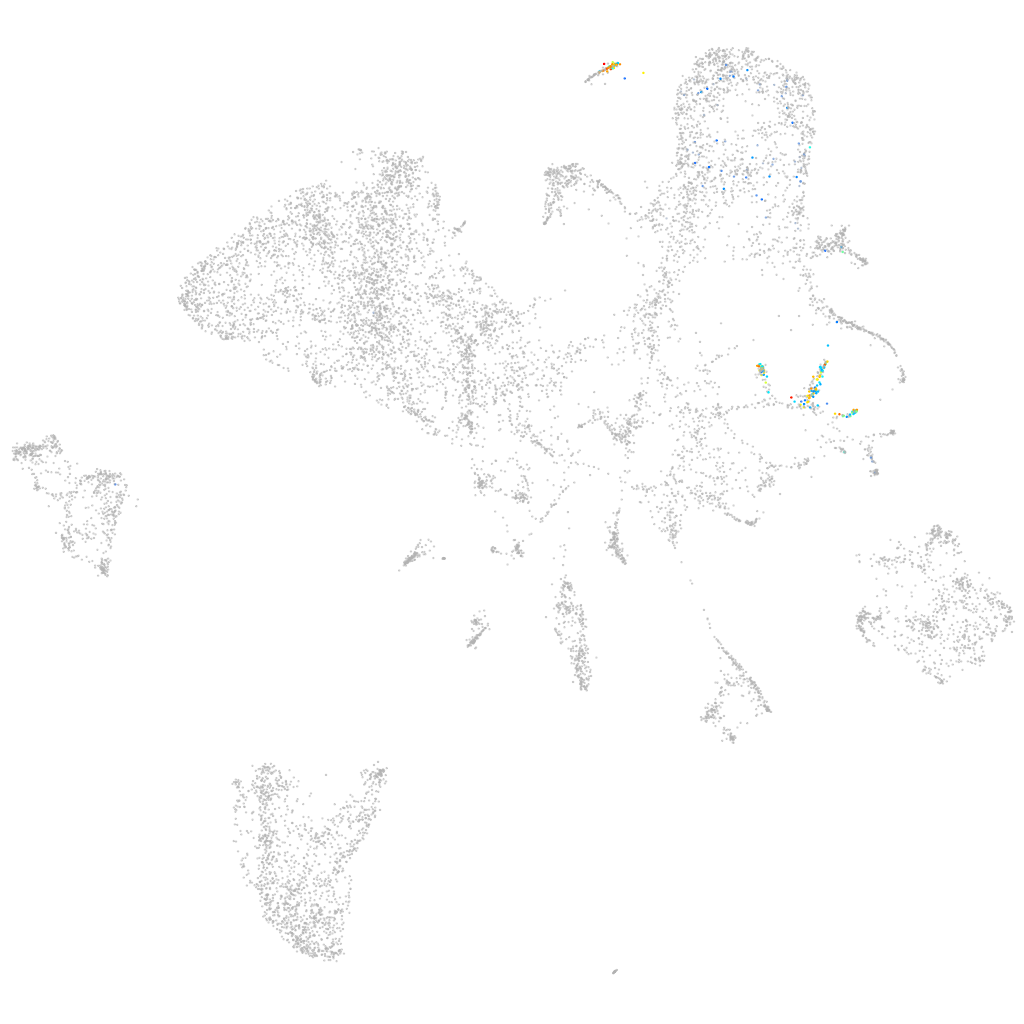
si:ch73-160i9.2
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.486 | ahcy | -0.129 |
| fev | 0.448 | fabp3 | -0.126 |
| c2cd4a | 0.447 | rpl6 | -0.113 |
| zgc:101731 | 0.445 | gamt | -0.107 |
| insm1b | 0.432 | rps20 | -0.105 |
| egr4 | 0.426 | hdlbpa | -0.104 |
| nmbb | 0.419 | gapdh | -0.104 |
| penka | 0.416 | bhmt | -0.104 |
| pcsk1nl | 0.403 | rps12 | -0.099 |
| TCIM (1 of many) | 0.403 | mat1a | -0.097 |
| alpk3b | 0.398 | aldh6a1 | -0.097 |
| neurod1 | 0.385 | gatm | -0.096 |
| arxa | 0.383 | eif4ebp1 | -0.096 |
| ccka | 0.369 | aqp12 | -0.096 |
| scgn | 0.359 | si:dkey-16p21.8 | -0.093 |
| hepacam2 | 0.359 | sec61a1 | -0.092 |
| mir7a-1 | 0.358 | apoc1 | -0.092 |
| rims2a | 0.354 | rpl4 | -0.091 |
| gfra3 | 0.353 | adka | -0.089 |
| gcga | 0.353 | agxtb | -0.089 |
| slc35g2a | 0.352 | agxta | -0.088 |
| tango2 | 0.349 | wu:fj16a03 | -0.087 |
| oprd1b | 0.345 | gstr | -0.087 |
| rprmb | 0.344 | shmt1 | -0.087 |
| syt1a | 0.343 | hspd1 | -0.086 |
| pcsk2 | 0.337 | gnmt | -0.086 |
| rasd4 | 0.337 | apoa4b.1 | -0.086 |
| capsla | 0.335 | fbp1b | -0.086 |
| vat1 | 0.333 | eif5a | -0.086 |
| pcsk1 | 0.331 | tfa | -0.086 |
| si:ch73-359m17.9 | 0.326 | cebpd | -0.086 |
| calm1b | 0.323 | gstt1a | -0.084 |
| BX088587.1 | 0.321 | apoa1b | -0.084 |
| capslb | 0.319 | eef2b | -0.084 |
| dll4 | 0.317 | cx32.3 | -0.083 |