si:ch211-282b22.1 [Source:ZFIN;Acc:ZDB-GENE-030219-148]
ZFIN 
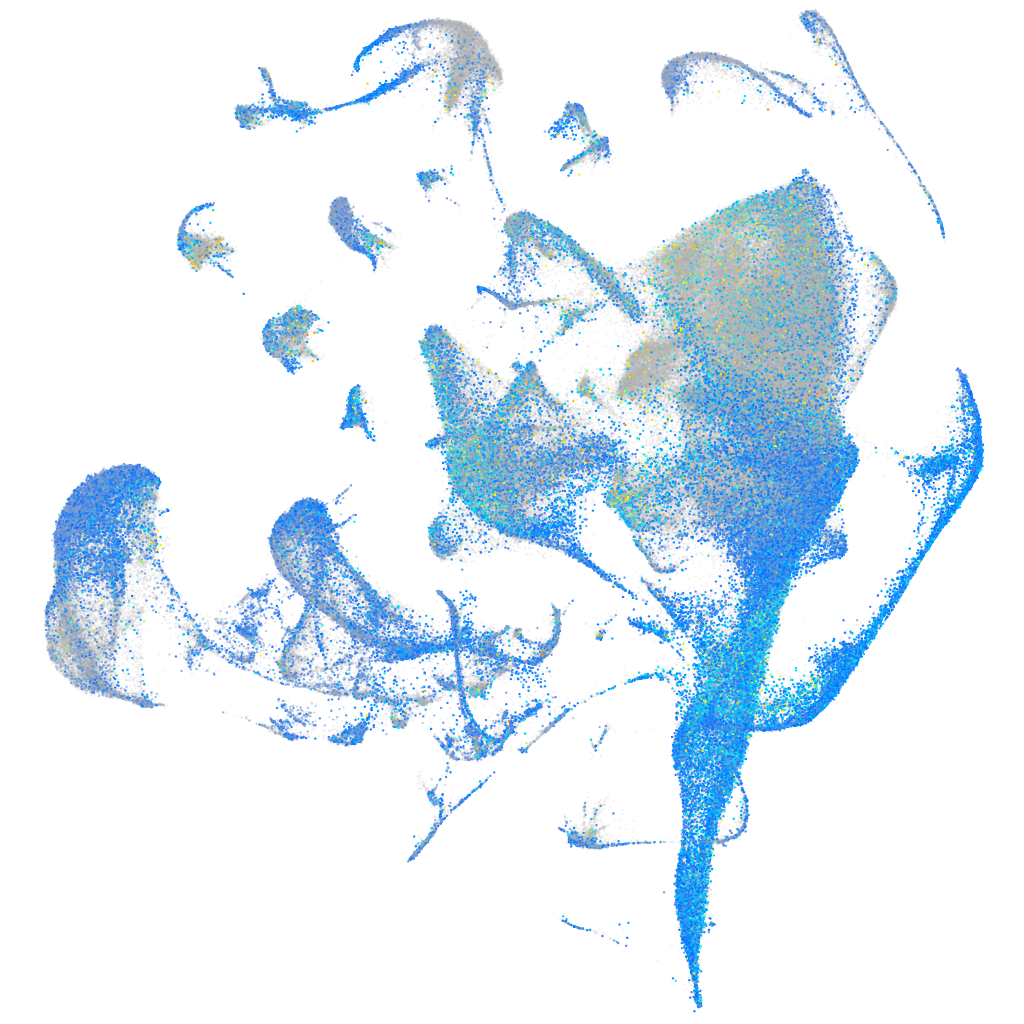
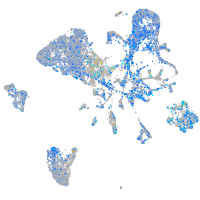
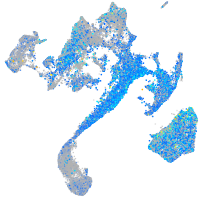
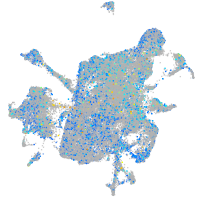
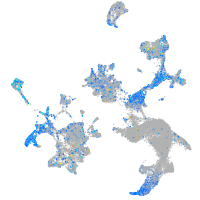
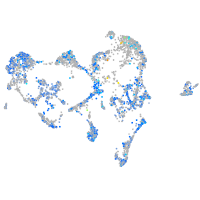
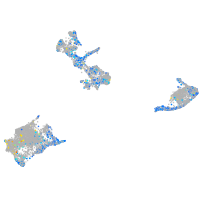
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| npm1a | 0.348 | tuba1c | -0.154 |
| snu13b | 0.313 | elavl3 | -0.148 |
| nop58 | 0.311 | gpm6aa | -0.148 |
| dkc1 | 0.307 | marcksl1a | -0.145 |
| fbl | 0.300 | rtn1a | -0.139 |
| nhp2 | 0.297 | nova2 | -0.138 |
| nop10 | 0.297 | stmn1b | -0.130 |
| nop56 | 0.285 | myt1b | -0.120 |
| nop2 | 0.284 | ckbb | -0.117 |
| ncl | 0.280 | gpm6ab | -0.115 |
| rsl1d1 | 0.279 | tuba1a | -0.115 |
| hspb1 | 0.276 | epb41a | -0.110 |
| rrp1 | 0.265 | zgc:153409 | -0.110 |
| si:ch211-217k17.7 | 0.263 | myt1a | -0.109 |
| ebna1bp2 | 0.262 | hbbe1.3 | -0.108 |
| pprc1 | 0.260 | tmsb | -0.106 |
| rpl7l1 | 0.258 | wu:fb55g09 | -0.106 |
| wdr43 | 0.256 | tubb5 | -0.104 |
| gnl3 | 0.253 | rnasekb | -0.102 |
| nip7 | 0.253 | CU634008.1 | -0.102 |
| mrto4 | 0.252 | ccni | -0.100 |
| gar1 | 0.251 | celf2 | -0.099 |
| c1qbp | 0.246 | LOC798783 | -0.099 |
| lyar | 0.245 | scrt2 | -0.099 |
| nifk | 0.243 | fabp3 | -0.098 |
| rrs1 | 0.242 | dpysl2b | -0.098 |
| bms1 | 0.241 | gng3 | -0.097 |
| mybbp1a | 0.239 | hbae3 | -0.097 |
| eif4ebp3l | 0.238 | fam168a | -0.096 |
| mak16 | 0.238 | insm1a | -0.095 |
| twistnb | 0.238 | pvalb1 | -0.095 |
| pes | 0.237 | gng2 | -0.093 |
| bxdc2 | 0.236 | pvalb2 | -0.093 |
| ssb | 0.235 | calm1a | -0.093 |
| ddx18 | 0.234 | CU467822.1 | -0.092 |