si:ch211-266o15.1
ZFIN 
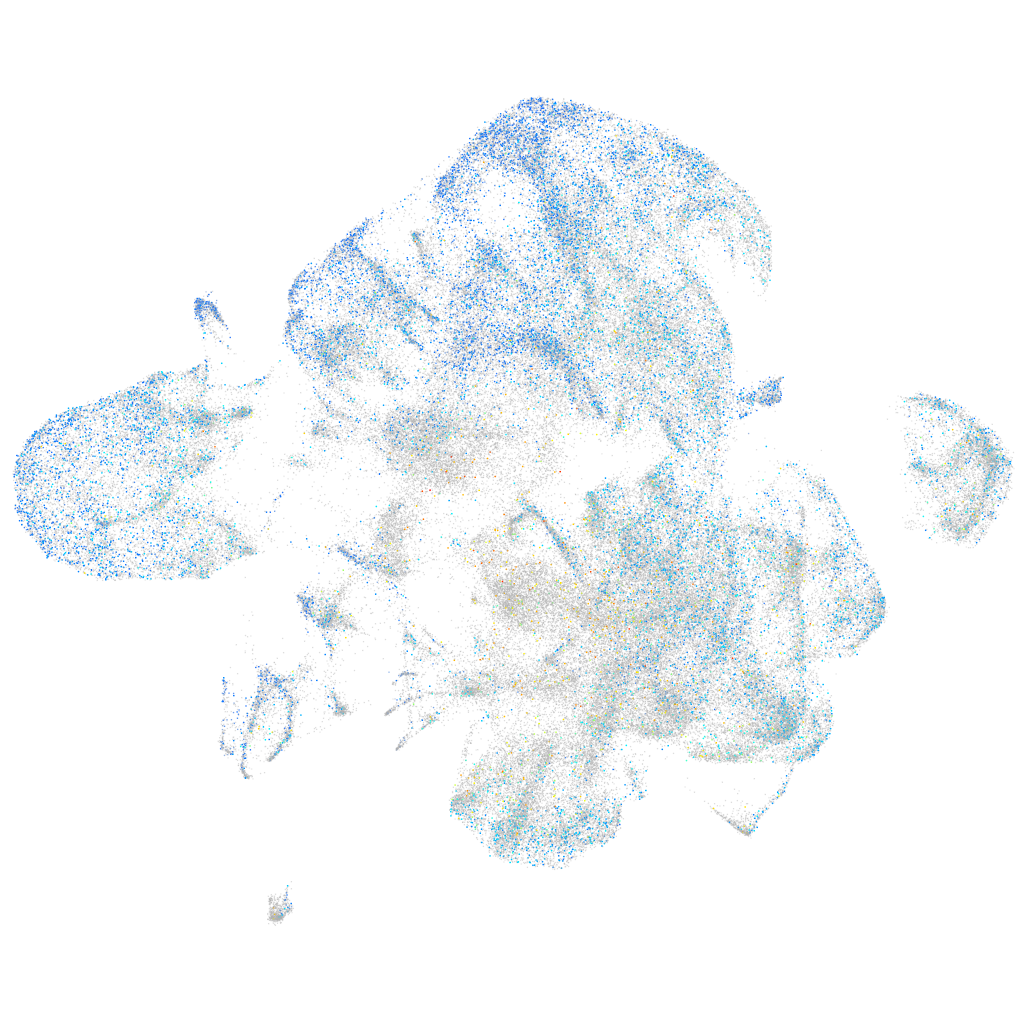
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mki67 | 0.069 | atp6v0cb | -0.043 |
| cirbpa | 0.067 | gapdhs | -0.041 |
| cx43.4 | 0.067 | pvalb1 | -0.037 |
| hmga1a | 0.066 | snap25a | -0.037 |
| khdrbs1a | 0.064 | rnasekb | -0.036 |
| ppm1g | 0.064 | sypb | -0.035 |
| seta | 0.062 | pvalb2 | -0.035 |
| snrpb | 0.062 | gpm6ab | -0.035 |
| ccnd1 | 0.062 | sncb | -0.035 |
| lig1 | 0.062 | sncgb | -0.035 |
| smc1al | 0.060 | si:dkeyp-75h12.5 | -0.033 |
| baz1b | 0.060 | nptna | -0.033 |
| fbxo5 | 0.060 | ptmaa | -0.033 |
| chaf1a | 0.060 | tpi1b | -0.033 |
| hnrnpa1b | 0.060 | atp6ap2 | -0.033 |
| msna | 0.059 | ywhag2 | -0.032 |
| eef2b | 0.059 | rtn1a | -0.032 |
| srsf1a | 0.059 | olfm1b | -0.032 |
| akap12b | 0.059 | scg2b | -0.032 |
| smarca4a | 0.058 | tmsb2 | -0.032 |
| chd7 | 0.058 | rtn1b | -0.031 |
| marcksb | 0.057 | eno2 | -0.031 |
| hnrnpub | 0.057 | ndufa4 | -0.031 |
| dek | 0.057 | calm1b | -0.031 |
| snrpd1 | 0.056 | actc1b | -0.030 |
| id1 | 0.056 | slc6a1a | -0.030 |
| syncrip | 0.056 | gng3 | -0.030 |
| hnrnpabb | 0.056 | map1aa | -0.030 |
| ddx39ab | 0.056 | cplx2 | -0.030 |
| sox19a | 0.056 | atpv0e2 | -0.029 |
| ccna2 | 0.055 | elavl4 | -0.029 |
| lbr | 0.055 | syt2a | -0.029 |
| cbx3a | 0.055 | stmn1b | -0.029 |
| hnrnpa0b | 0.055 | aplp1 | -0.029 |
| eif5a2 | 0.055 | ckbb | -0.029 |