si:ch211-255i20.3
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural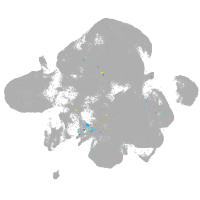 |
spinal cord / glia |
eye |
taste / olfactory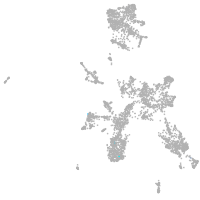 |
otic / lateral line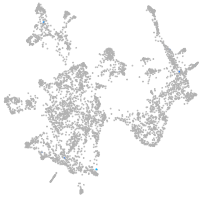 |
epidermis |
periderm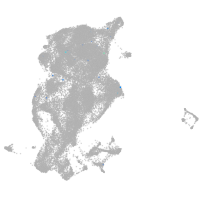 |
fin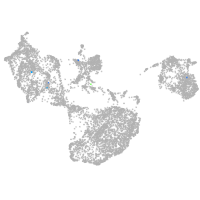 |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CU207241.1 | 0.668 | rpl6 | -0.055 |
| tradv10.0.5 | 0.642 | epcam | -0.047 |
| LOC110438072 | 0.637 | cldnb | -0.042 |
| si:ch211-194m7.3 | 0.589 | hsp90ab1 | -0.039 |
| gpc5b | 0.533 | ubb | -0.039 |
| si:ch73-352p4.8 | 0.489 | btg1 | -0.039 |
| cpa4 | 0.439 | cnbpb | -0.036 |
| msrb2 | 0.432 | rps3 | -0.036 |
| vipb | 0.431 | rpl30 | -0.035 |
| capn3b | 0.431 | atp5f1e | -0.035 |
| hspb15 | 0.411 | atp5mc3a | -0.035 |
| bin2a | 0.363 | mibp2 | -0.035 |
| fabp10b | 0.358 | eif2s2 | -0.035 |
| apba2a | 0.355 | mt-co2 | -0.033 |
| cacna1da | 0.282 | hnrnpa0l | -0.033 |
| celsr1b | 0.275 | si:ch211-196l7.4 | -0.032 |
| avpr2ab | 0.272 | eif3g | -0.032 |
| apoba | 0.254 | rps18 | -0.032 |
| ntn1b | 0.251 | serbp1a | -0.032 |
| npas1 | 0.247 | ndufs7 | -0.031 |
| kirrel1a | 0.242 | oclna | -0.031 |
| kif5aa | 0.238 | glrx | -0.031 |
| s1pr1 | 0.225 | eif2s3 | -0.031 |
| jam3b | 0.222 | cox8a | -0.030 |
| LOC110437731 | 0.219 | atp5pd | -0.030 |
| CABZ01081909.1 | 0.215 | cox5aa | -0.030 |
| ptgdsb.2 | 0.211 | cct6a | -0.030 |
| adra2a | 0.210 | gng5 | -0.030 |
| slc29a1a | 0.208 | ywhae1 | -0.030 |
| aldh3b2 | 0.199 | ier5 | -0.030 |
| cd81b | 0.197 | mt-nd2 | -0.030 |
| epor | 0.196 | gtpbp4 | -0.029 |
| phyh | 0.191 | si:dkey-112e7.2 | -0.029 |
| ndrg1b | 0.175 | psmb4 | -0.029 |
| clip2 | 0.171 | syncrip | -0.029 |