si:ch211-226l4.6
ZFIN 
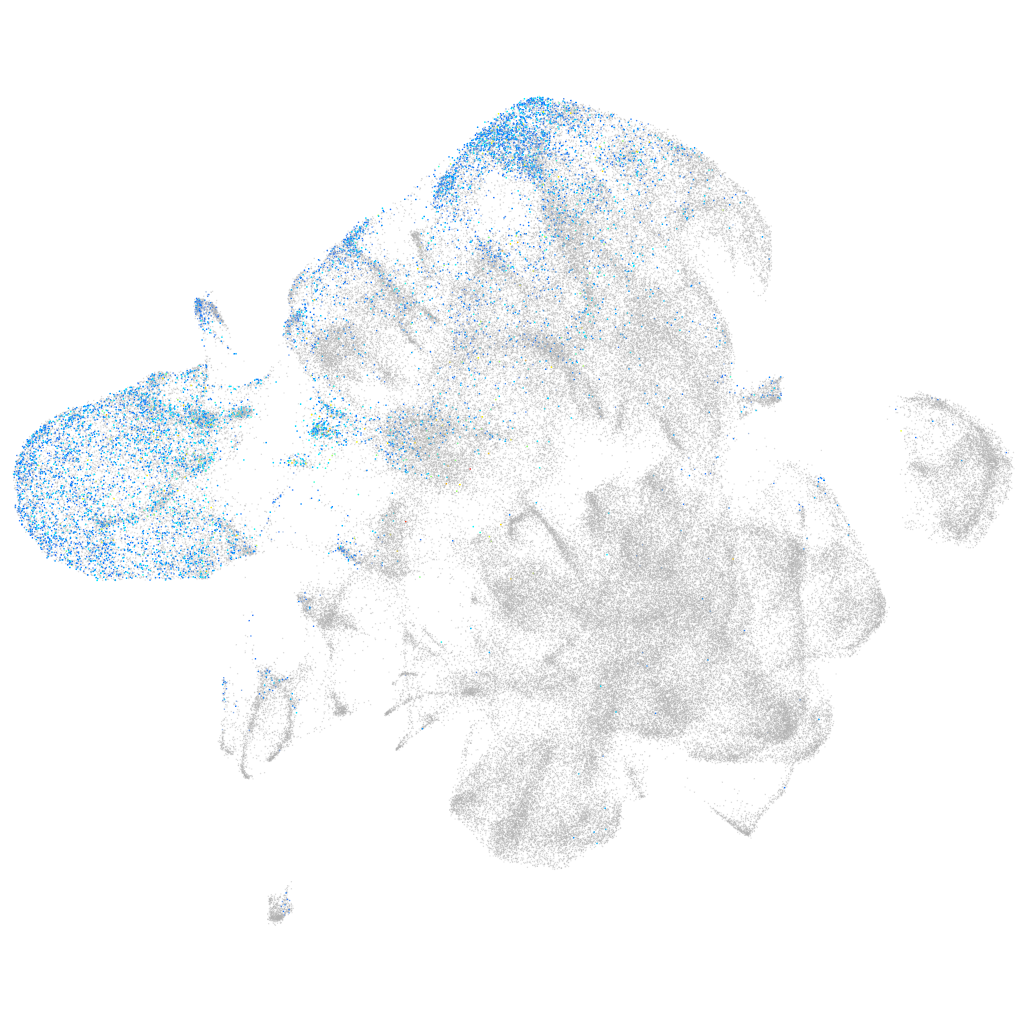
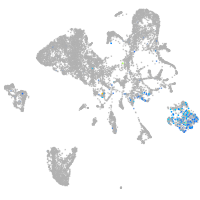
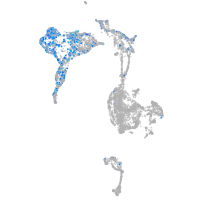
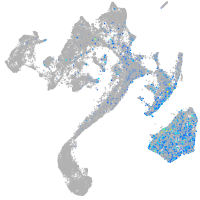
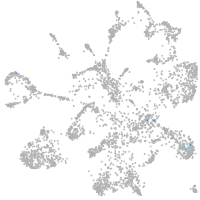
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hspb1 | 0.349 | ptmaa | -0.225 |
| npm1a | 0.295 | CR383676.1 | -0.212 |
| nop58 | 0.284 | rtn1a | -0.208 |
| BX927258.1 | 0.282 | elavl3 | -0.206 |
| dkc1 | 0.275 | gpm6aa | -0.203 |
| fbl | 0.273 | tuba1c | -0.199 |
| si:ch211-152c2.3 | 0.267 | stmn1b | -0.197 |
| snu13b | 0.260 | ckbb | -0.180 |
| nop56 | 0.260 | tmsb | -0.179 |
| lig1 | 0.253 | COX3 | -0.172 |
| XLOC-032526 | 0.250 | gpm6ab | -0.170 |
| nop2 | 0.249 | marcksl1a | -0.164 |
| apoeb | 0.249 | gng3 | -0.158 |
| nhp2 | 0.247 | rnasekb | -0.156 |
| anp32b | 0.245 | pvalb1 | -0.154 |
| aldob | 0.244 | nova2 | -0.154 |
| apela | 0.243 | celf2 | -0.153 |
| rsl1d1 | 0.242 | tmsb4x | -0.153 |
| ncl | 0.240 | sncb | -0.151 |
| gar1 | 0.237 | pvalb2 | -0.151 |
| banf1 | 0.236 | rpl37 | -0.150 |
| mcm6 | 0.236 | atp6v0cb | -0.150 |
| ebna1bp2 | 0.235 | gapdhs | -0.147 |
| nop10 | 0.234 | rtn1b | -0.144 |
| stm | 0.233 | zgc:158463 | -0.144 |
| chaf1a | 0.228 | ccni | -0.144 |
| pcna | 0.227 | calm1a | -0.143 |
| mcm3 | 0.226 | actc1b | -0.143 |
| s100a1 | 0.226 | ppiab | -0.142 |
| hmgb2a | 0.226 | elavl4 | -0.142 |
| lyar | 0.225 | mt-co2 | -0.141 |
| nr6a1a | 0.225 | ppdpfb | -0.141 |
| nasp | 0.224 | ywhah | -0.139 |
| ranbp1 | 0.221 | vamp2 | -0.138 |
| gnl3 | 0.219 | zgc:114188 | -0.138 |