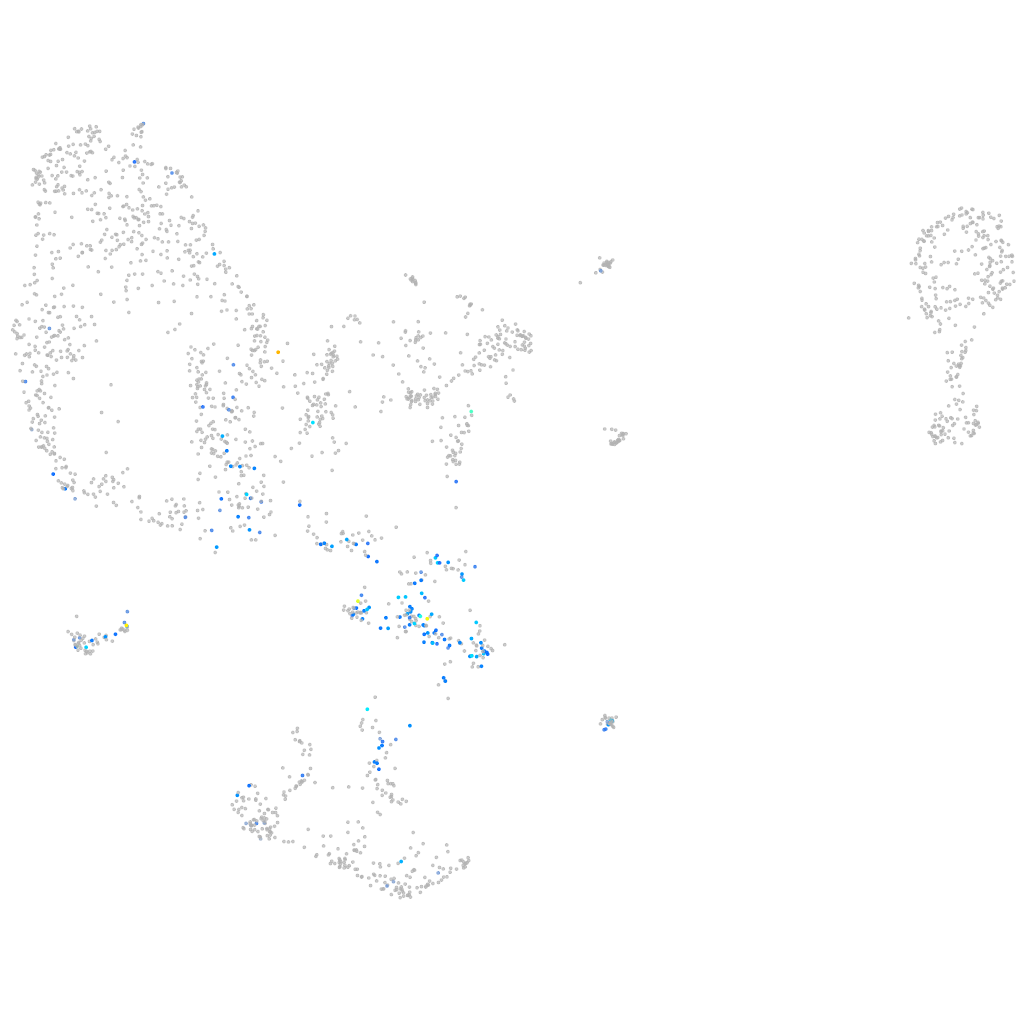
si:ch211-220f16.1
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ccdc151 | 0.396 | si:ch211-139a5.9 | -0.199 |
| foxj1a | 0.374 | gapdh | -0.175 |
| CR388369.1 | 0.374 | si:dkey-16p21.8 | -0.163 |
| BX469925.3 | 0.373 | rplp2 | -0.159 |
| vcana | 0.370 | gcshb | -0.154 |
| ribc2 | 0.358 | glud1b | -0.153 |
| rab36 | 0.352 | aldh6a1 | -0.150 |
| si:dkey-114c15.5 | 0.350 | mt-nd3 | -0.150 |
| dnaaf4 | 0.328 | pdzk1ip1 | -0.149 |
| dpcd | 0.326 | gstt1a | -0.149 |
| cfap20 | 0.325 | aldob | -0.148 |
| flvcr1 | 0.324 | si:dkey-33i11.9 | -0.146 |
| miip | 0.322 | eno3 | -0.145 |
| cd151l | 0.321 | ahcy | -0.144 |
| CT025771.1 | 0.321 | cox6a2 | -0.143 |
| tmem17 | 0.321 | cubn | -0.143 |
| cfap45 | 0.320 | hao1 | -0.142 |
| rabl2 | 0.316 | alpl | -0.140 |
| cfap298 | 0.316 | prdx2 | -0.139 |
| pih1d2 | 0.315 | cdaa | -0.138 |
| ccdc173 | 0.315 | grhprb | -0.137 |
| daw1 | 0.313 | phyhd1 | -0.135 |
| ncs1a | 0.309 | ldhba | -0.135 |
| efcab2 | 0.306 | rps18 | -0.134 |
| dnal1 | 0.305 | haao | -0.133 |
| si:ch211-163l21.7 | 0.303 | dpys | -0.133 |
| ift57 | 0.300 | agxtb | -0.133 |
| stk33 | 0.298 | sod1 | -0.132 |
| e2f5 | 0.297 | gstr | -0.132 |
| si:dkey-42p14.3 | 0.296 | gstp1 | -0.131 |
| rpp25a | 0.292 | cox5b2 | -0.131 |
| XLOC-001964 | 0.292 | hoga1 | -0.130 |
| tpm4a | 0.292 | sod2 | -0.130 |
| ftr82 | 0.289 | zgc:85777 | -0.129 |
| spag6 | 0.288 | cox8b | -0.129 |