si:ch211-210b2.1
ZFIN 
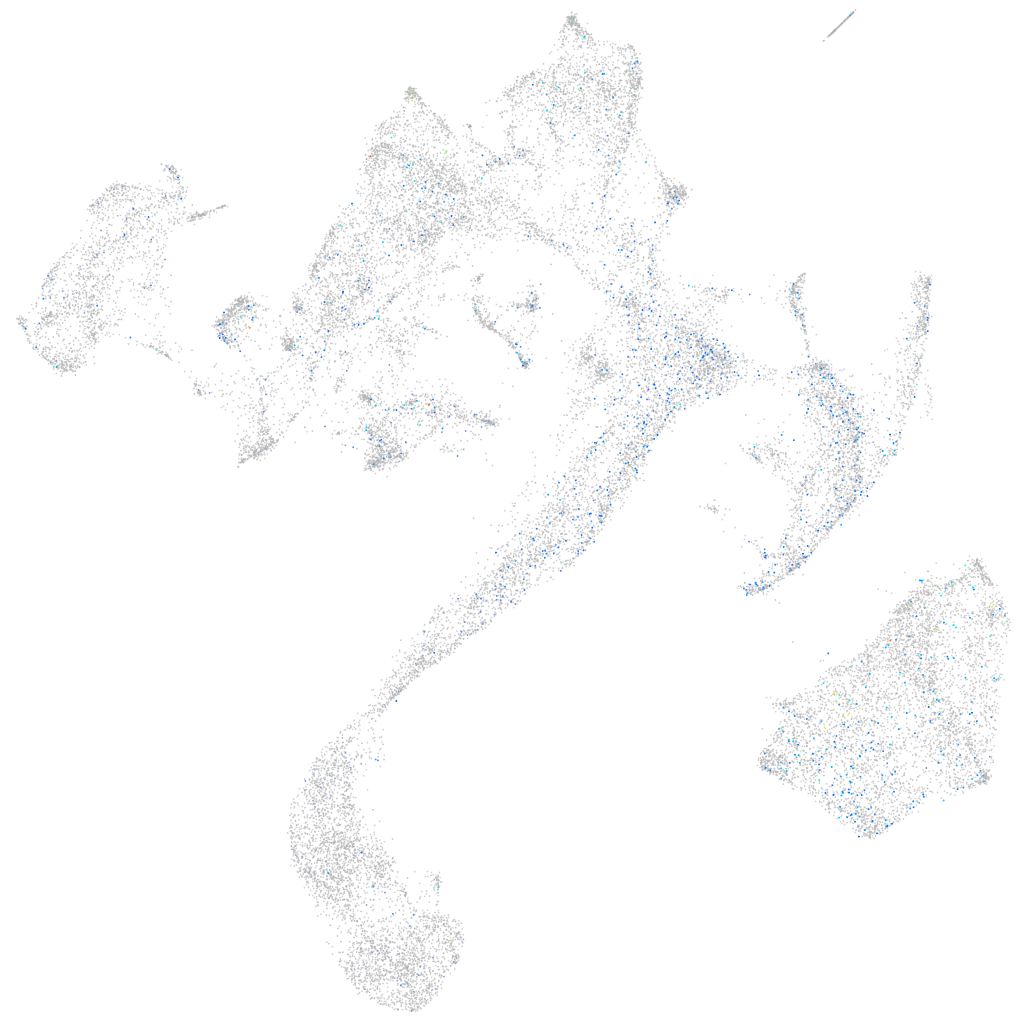
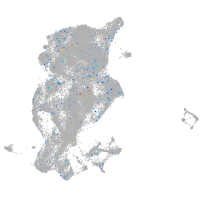
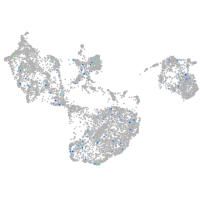
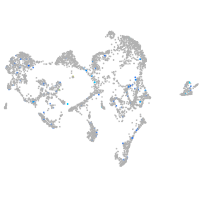
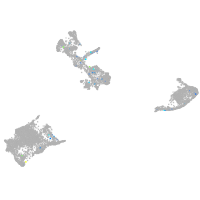
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc22a4 | 0.092 | actc1b | -0.065 |
| nop56 | 0.081 | ak1 | -0.062 |
| npm1a | 0.080 | ckma | -0.061 |
| setb | 0.079 | tnnc2 | -0.059 |
| nop58 | 0.078 | ckmb | -0.059 |
| ncl | 0.077 | atp2a1 | -0.057 |
| ptges3b | 0.077 | actn3a | -0.056 |
| khdrbs1a | 0.076 | acta1b | -0.056 |
| fbl | 0.075 | si:ch73-367p23.2 | -0.056 |
| snu13b | 0.075 | tpma | -0.056 |
| syncrip | 0.075 | bhmt | -0.056 |
| hnrnpabb | 0.075 | mylpfa | -0.055 |
| snrpd2 | 0.073 | aldoab | -0.055 |
| hnrnpaba | 0.073 | neb | -0.055 |
| srsf5a | 0.072 | ldb3b | -0.055 |
| snrpb | 0.072 | actn3b | -0.055 |
| ddx39ab | 0.071 | myom1a | -0.055 |
| ebna1bp2 | 0.071 | mylz3 | -0.055 |
| cirbpa | 0.071 | tnnt3a | -0.055 |
| nop2 | 0.071 | smyd1a | -0.054 |
| hmga1a | 0.071 | si:dkey-16p21.8 | -0.054 |
| nhp2 | 0.070 | eno3 | -0.053 |
| nop10 | 0.069 | eno1a | -0.053 |
| dkc1 | 0.069 | pvalb2 | -0.053 |
| pprc1 | 0.068 | tmod4 | -0.053 |
| srsf11 | 0.068 | tpi1b | -0.053 |
| cbx3a | 0.068 | nme2b.2 | -0.052 |
| h2afvb | 0.068 | CABZ01078594.1 | -0.052 |
| cx43.4 | 0.068 | casq2 | -0.052 |
| pabpc1a | 0.068 | pvalb1 | -0.052 |
| magoh | 0.068 | pgam2 | -0.052 |
| si:ch73-281n10.2 | 0.068 | tmem38a | -0.052 |
| rrp1 | 0.067 | si:ch211-266g18.10 | -0.051 |
| cnbpa | 0.067 | ank1a | -0.051 |
| seta | 0.067 | gapdh | -0.051 |