si:ch211-193k19.2
ZFIN 
Other cell groups


all cells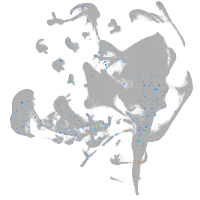 |
blastomeres |
PGCs |
endoderm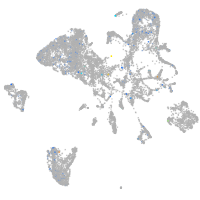 |
axial mesoderm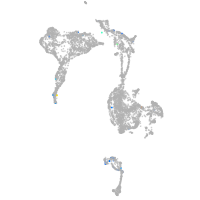 |
muscle 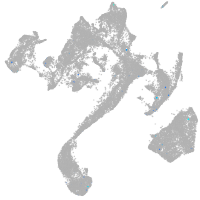 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 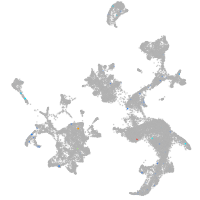 |
pronephros 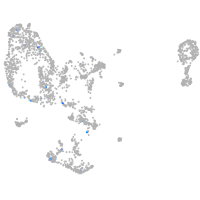 |
neural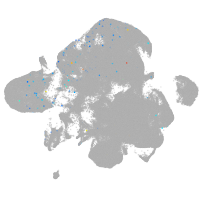 |
spinal cord / glia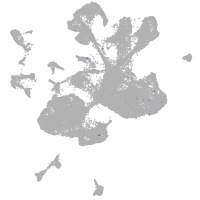 |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm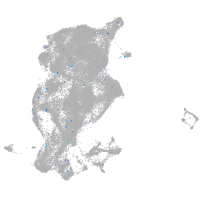 |
fin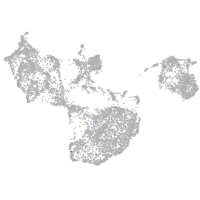 |
ionocytes / mucous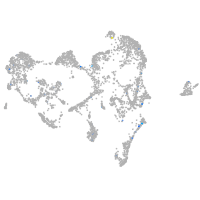 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:171242 | 0.047 | gpm6aa | -0.029 |
| cx32.3 | 0.046 | marcksl1a | -0.027 |
| rltgr | 0.045 | gpm6ab | -0.026 |
| rdh1 | 0.043 | stmn1b | -0.026 |
| CR855996.2 | 0.042 | ckbb | -0.025 |
| tm4sf5 | 0.042 | elavl3 | -0.025 |
| creb3l3a | 0.041 | hmgb3a | -0.024 |
| fbp1b | 0.041 | nova2 | -0.023 |
| st3gal7 | 0.041 | celf2 | -0.022 |
| clic5b | 0.041 | tuba1c | -0.022 |
| cx28.9 | 0.041 | actc1b | -0.021 |
| slc37a4a | 0.040 | gng3 | -0.021 |
| ugt1a7 | 0.040 | rtn1a | -0.021 |
| si:ch211-248a14.8 | 0.040 | tmsb | -0.021 |
| aqp12 | 0.039 | hmgb1b | -0.020 |
| cyp8b1 | 0.039 | jpt1b | -0.020 |
| ezra | 0.039 | zc4h2 | -0.020 |
| gsta.1 | 0.039 | fez1 | -0.019 |
| krt92 | 0.039 | fscn1a | -0.019 |
| si:dkey-16i5.8 | 0.039 | myt1b | -0.019 |
| cideb | 0.038 | pvalb2 | -0.019 |
| cox8b | 0.038 | si:dkey-56m19.5 | -0.019 |
| cyp2p9 | 0.038 | tubb5 | -0.019 |
| golt1a | 0.038 | cspg5a | -0.018 |
| pla2g12b | 0.038 | CU467822.1 | -0.018 |
| pnp4b | 0.038 | elavl4 | -0.018 |
| CU682777.2 | 0.037 | hnrnpa0a | -0.018 |
| cyp4v8 | 0.037 | myt1a | -0.018 |
| glod5 | 0.037 | pvalb1 | -0.018 |
| mttp | 0.037 | rtn1b | -0.018 |
| pbld | 0.037 | scrt2 | -0.018 |
| si:ch211-117n7.7 | 0.037 | si:ch211-133n4.4 | -0.018 |
| ugt5b3 | 0.037 | si:dkey-276j7.1 | -0.018 |
| acmsd | 0.036 | si:dkeyp-75h12.5 | -0.018 |
| apoa4b.1 | 0.036 | CU634008.1 | -0.018 |




