si:ch211-154o6.2
ZFIN 
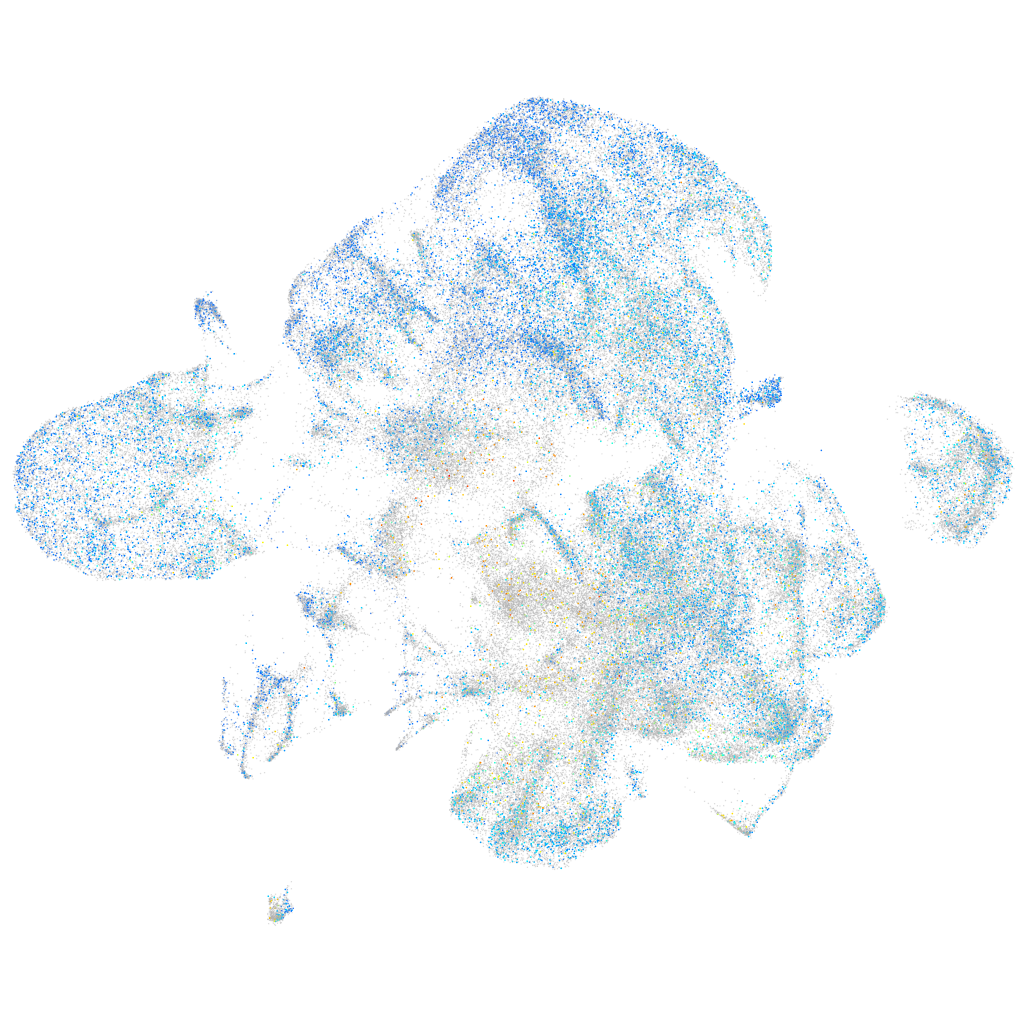
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpaba | 0.073 | apoa2 | -0.032 |
| khdrbs1a | 0.068 | apoa1b | -0.030 |
| si:ch211-288g17.3 | 0.066 | pvalb2 | -0.029 |
| hdac1 | 0.066 | prss1 | -0.027 |
| sumo3a | 0.063 | grin1b | -0.026 |
| ptmab | 0.063 | pvalb1 | -0.025 |
| hnrnpabb | 0.062 | CELA1 (1 of many) | -0.023 |
| si:ch211-222l21.1 | 0.062 | prss59.2 | -0.023 |
| hnrnpa0b | 0.061 | ela2l | -0.023 |
| cirbpb | 0.061 | slc6a1a | -0.022 |
| h2afvb | 0.058 | ctrb1 | -0.022 |
| hmgn6 | 0.057 | edil3a | -0.022 |
| tubb2b | 0.057 | fxyd6 | -0.022 |
| hmgb1b | 0.057 | atp2b3b | -0.022 |
| h2afva | 0.057 | sncgb | -0.022 |
| hmga1a | 0.056 | si:dkey-7j14.5 | -0.022 |
| h3f3d | 0.056 | elmod1 | -0.022 |
| ddx39ab | 0.054 | nptna | -0.021 |
| setb | 0.053 | actc1b | -0.021 |
| ybx1 | 0.053 | BX005003.1 | -0.021 |
| cirbpa | 0.053 | fabp10a | -0.021 |
| hmgb2b | 0.053 | adgrl3.1 | -0.021 |
| ubb | 0.052 | glula | -0.020 |
| syncrip | 0.052 | COX3 | -0.020 |
| snrpd2 | 0.052 | syngr1a | -0.020 |
| alyref | 0.052 | lgals2b | -0.020 |
| hmgn2 | 0.052 | sypb | -0.020 |
| psmb7 | 0.052 | scn2b | -0.020 |
| cbx3a | 0.052 | scg2b | -0.020 |
| ran | 0.051 | mylz3 | -0.020 |
| hnrnph1l | 0.051 | slc6a17 | -0.020 |
| seta | 0.051 | IGLON5 | -0.020 |
| tpt1 | 0.051 | sypa | -0.020 |
| erh | 0.051 | ela2 | -0.019 |
| nono | 0.050 | oaz2b | -0.019 |