si:ch211-137a8.4
ZFIN 
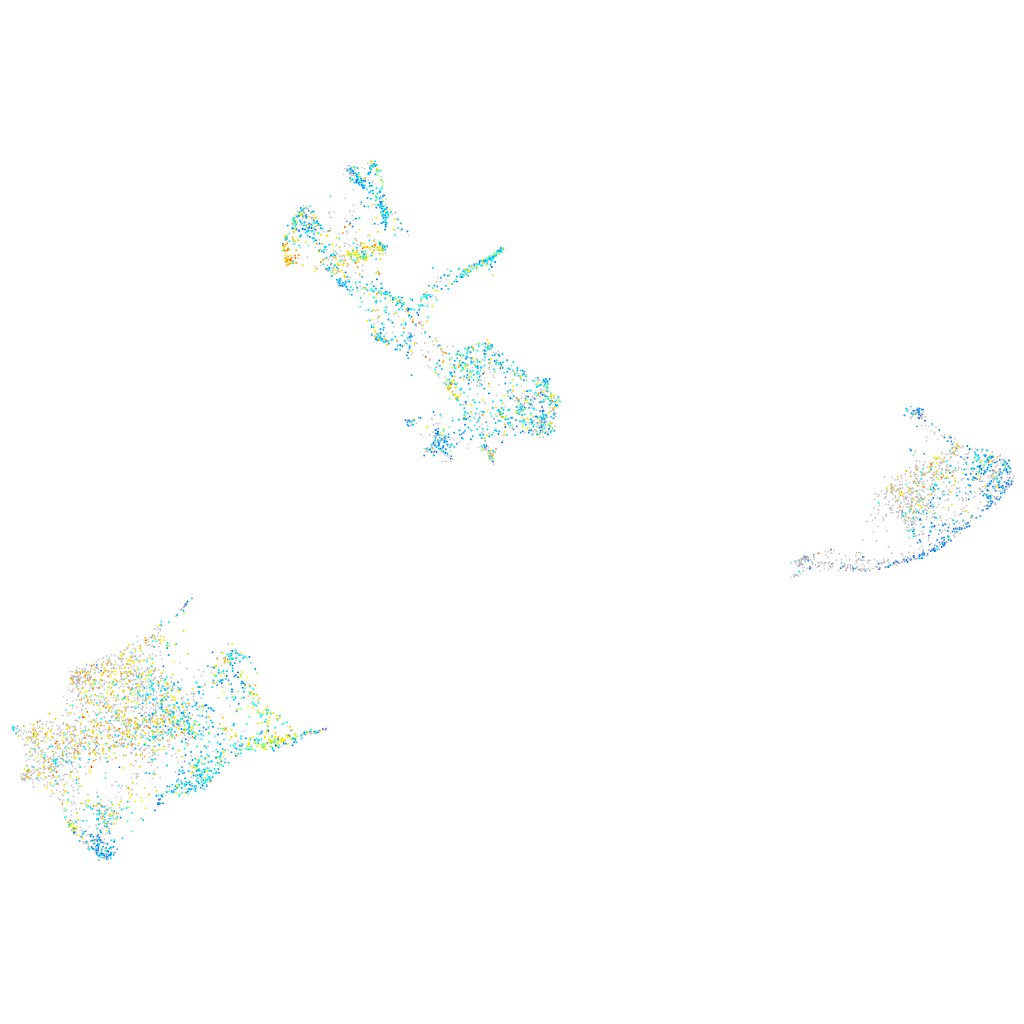
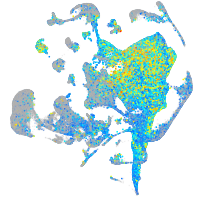
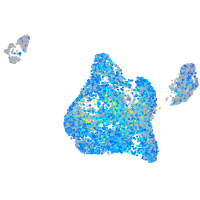
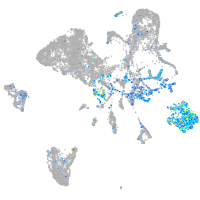
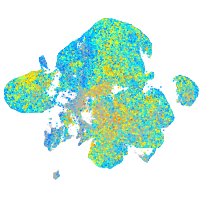
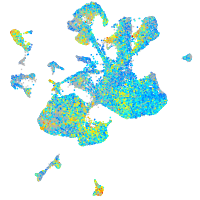
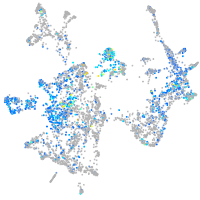
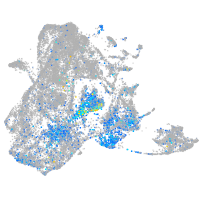
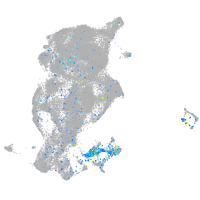
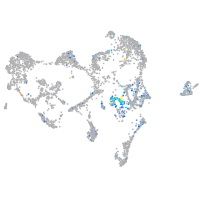
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gmps | 0.224 | tyrp1b | -0.199 |
| impdh1b | 0.211 | tyrp1a | -0.199 |
| glulb | 0.202 | pmela | -0.190 |
| guk1a | 0.193 | dct | -0.190 |
| CABZ01072077.1 | 0.192 | zgc:91968 | -0.175 |
| defbl1 | 0.191 | oca2 | -0.172 |
| pltp | 0.191 | si:ch73-389b16.1 | -0.167 |
| apoda.1 | 0.190 | slc24a5 | -0.159 |
| atic | 0.190 | tyr | -0.148 |
| ppat | 0.187 | anxa1a | -0.147 |
| slc25a36a | 0.187 | icn | -0.147 |
| si:ch73-89b15.3 | 0.186 | kita | -0.147 |
| adsl | 0.186 | s100a10b | -0.146 |
| unm-sa821 | 0.184 | mtbl | -0.138 |
| si:ch211-256m1.8 | 0.184 | gstt1a | -0.135 |
| slc2a15a | 0.183 | prkar1b | -0.130 |
| pax7a | 0.181 | aadac | -0.124 |
| gpnmb | 0.180 | mlpha | -0.122 |
| paics | 0.180 | slc29a3 | -0.117 |
| alx1 | 0.178 | tmem243b | -0.113 |
| aqp1a.1 | 0.177 | qdpra | -0.108 |
| si:dkey-197i20.6 | 0.176 | slc45a2 | -0.107 |
| alx4a | 0.176 | tfap2e | -0.105 |
| prps1a | 0.174 | cd63 | -0.105 |
| tuba1b | 0.170 | pcbd1 | -0.104 |
| ttc39a | 0.170 | slc7a5 | -0.104 |
| pnp4a | 0.170 | slc22a2 | -0.098 |
| tfec | 0.169 | csrp2 | -0.098 |
| lypc | 0.167 | lamp1a | -0.098 |
| akap12a | 0.167 | slc39a10 | -0.096 |
| dhrs3b | 0.167 | mical3b | -0.096 |
| npffr1l2 | 0.163 | kcnj13 | -0.095 |
| sdc2 | 0.162 | rap1gap | -0.095 |
| tcirg1a | 0.162 | pknox1.2 | -0.094 |
| shmt1 | 0.162 | BX248318.1 | -0.089 |