si:ch211-114n24.6
ZFIN 
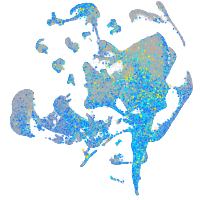
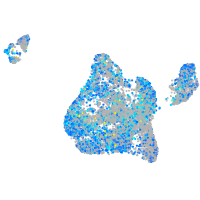
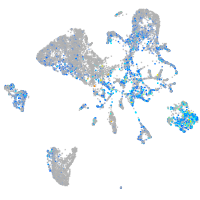
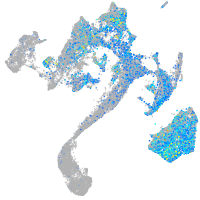
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rrm1 | 0.308 | CR383676.1 | -0.158 |
| hmgb2a | 0.300 | gpm6ab | -0.150 |
| ccna2 | 0.299 | ckbb | -0.145 |
| pcna | 0.292 | rtn1a | -0.140 |
| tuba8l4 | 0.291 | elavl4 | -0.138 |
| dut | 0.291 | ppdpfb | -0.138 |
| mki67 | 0.288 | atp6v0cb | -0.134 |
| cks1b | 0.285 | rnasekb | -0.134 |
| lbr | 0.280 | stx1b | -0.132 |
| rrm2 | 0.277 | gng3 | -0.130 |
| tuba8l | 0.270 | stxbp1a | -0.128 |
| stmn1a | 0.267 | calm1a | -0.128 |
| chaf1a | 0.267 | si:dkeyp-75h12.5 | -0.127 |
| ranbp1 | 0.266 | sncb | -0.127 |
| dek | 0.265 | elavl3 | -0.126 |
| banf1 | 0.262 | stmn1b | -0.126 |
| zgc:110216 | 0.262 | snap25a | -0.125 |
| anp32b | 0.261 | vamp2 | -0.125 |
| hmgb2b | 0.256 | myt1b | -0.124 |
| mad2l1 | 0.256 | cplx2 | -0.120 |
| aurkb | 0.255 | atp6v1e1b | -0.119 |
| smc2 | 0.255 | gabarapl2 | -0.116 |
| cdca5 | 0.250 | cadm4 | -0.116 |
| mibp | 0.249 | gapdhs | -0.115 |
| snrpd1 | 0.248 | sncgb | -0.115 |
| tpx2 | 0.248 | syt2a | -0.113 |
| seta | 0.247 | acbd7 | -0.112 |
| ncapg | 0.247 | hbae3 | -0.111 |
| hmga1a | 0.245 | aldocb | -0.110 |
| cbx3a | 0.244 | atpv0e2 | -0.110 |
| nasp | 0.242 | aplp1 | -0.109 |
| hmgn2 | 0.241 | rbfox1 | -0.108 |
| snrpf | 0.241 | map1aa | -0.108 |
| mcm7 | 0.238 | ywhag2 | -0.107 |
| rpa2 | 0.238 | fez1 | -0.106 |